Single-cell cloning (SCC)
Once the vector carrying the product transgene is integrated into the CHO genome, SCC is performed to confirm stable production and high protein quality. SCC is a critical step in cell line to development. It ensures that the selected clones reliably produce the protein of interest with appropriate PTMs.
For scientists who need to screen hundreds or thousands of clones, manual screening methods are tedious, labor-intensive, and time-consuming. The massive project scale underscores the need for high-throughput solutions to process stacks of plates or dozens of flasks.
The goals of SCC are to:
- Prevent overgrowth of low producers
- Promote stable survival of high producers
Achieving these goals requires screening THOUSANDS of clones over a period of months.
Nexcelom platforms provide rapid, high-throughput imaging and analysis with the ability to document monoclonality and expansion with ease.
Clonality is one of the most crucial steps in guaranteeing cell line quality and safety. The FDA does not currently specify the acceptable percentage of clonality or which single-cell sorting methods are recommended, but they do require evidence that each cell line used in manufacturing has been generated from a single cell. Celigo provides image-based proof of single-cell clonality.
Single cell cloning (SCC) for cell line development
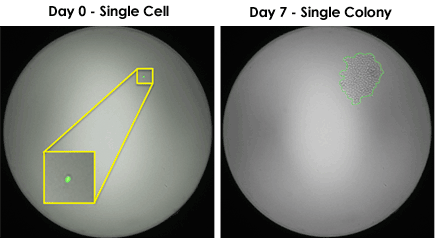
Monoclonality starts with identifying single-cell located in each well, which can be achieved by two common methods. (1) Single-cell sorting using FACS and (2) limiting dilution. Specifically for limiting dilution, prior to the process, it is important to obtain accurate cell counting results in order to have proper dilution into the plates.
Clonal validation on Celigo provides several benefits:
- Rapid brightfield scanning of multi-well plates (384-well plate in <5 min)
- Accurate segmentation for detection of single cells
- Heatmap representation of the number of cells/well
- Growth tracking reporting for monitoring over time
- Efficient database management of images and data
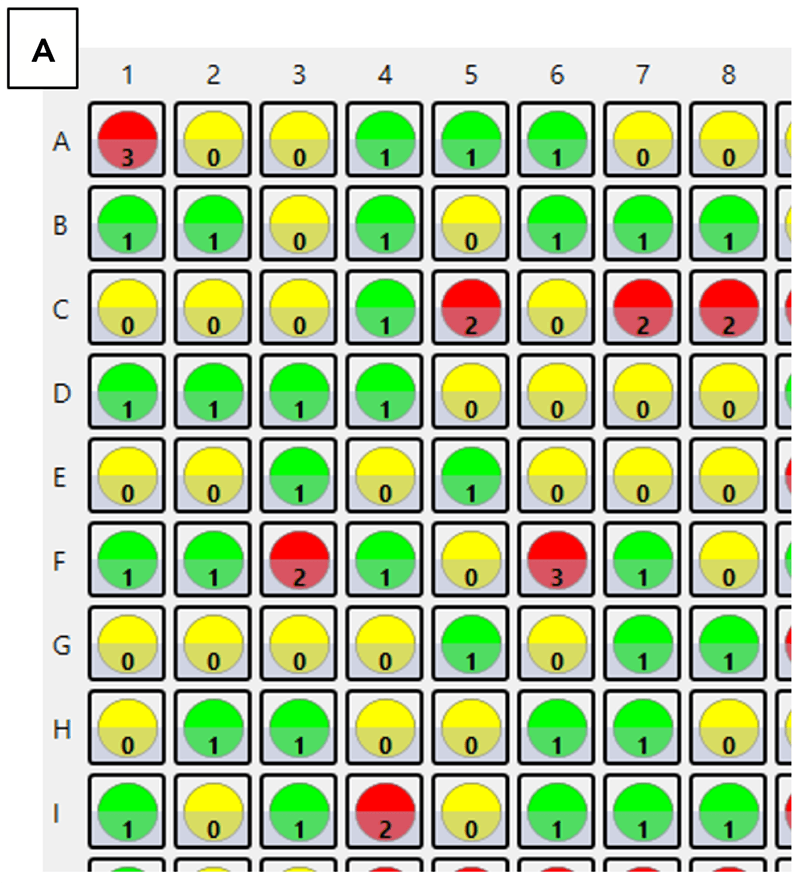
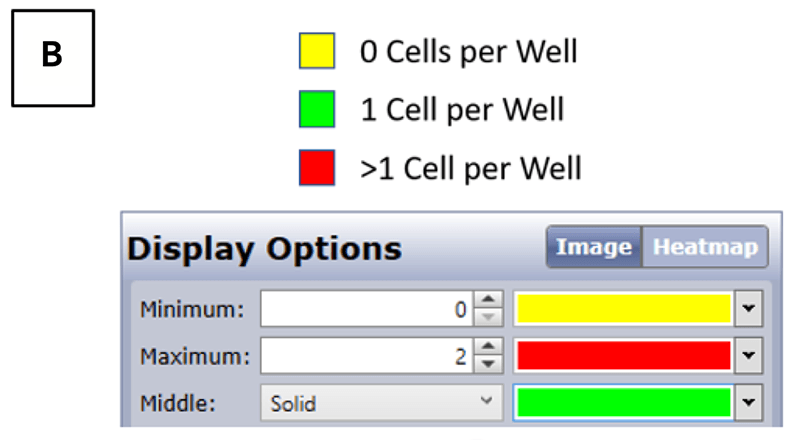
Heatmaps generated by the Celigo Image Cytometer after single-cell plating. (A) Reported cell counts following a limiting dilution protocol in 384-well Corning 3542 plates. (B) Heatmap legends and user-defined settings for identifying 1 cell per well.
Celigo Image Cytometer rapidly identifies single cells in 96- or 384-well plates
In 2018 Zigon et al. conducted a collaborative project with Nexcelom Bioscience and Beth Israel Deaconess Medical Center Flow Cytometry Core to develop a high-throughput single cell detection method using the Celigo Image Cytometer. Jurkat cells were stained with calcein AM (green), single cell sorted into a 96-well plate, and imaged and analyzed by the Celigo. In addition to images, the platform provides an easy-to-understand map indicating which wells were seeded with multiple cells.
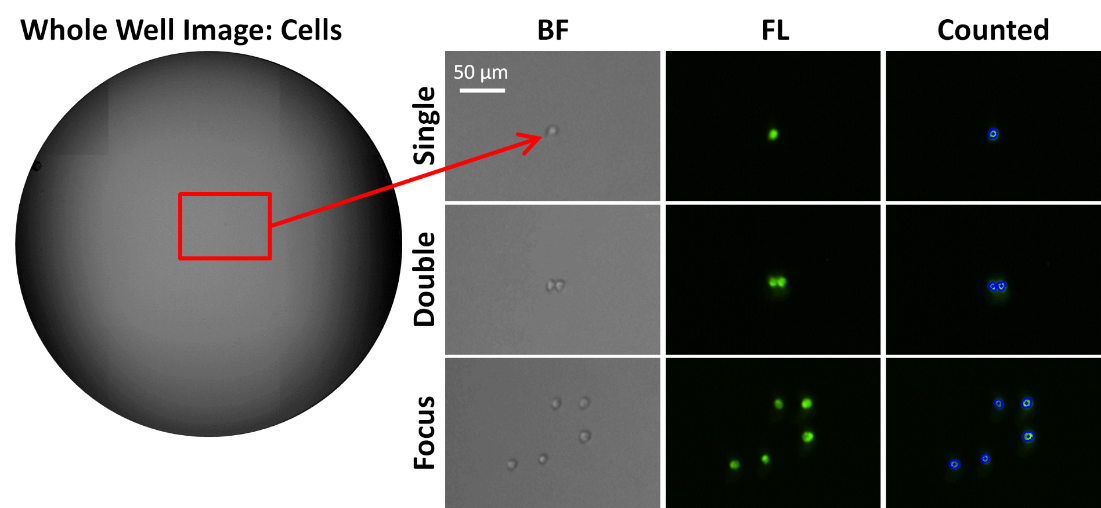

In addition, the Early Stage Cell Culture group at Genentech, Inc. demonstrated high single-cell detection accuracy with the Celigo for cells stained with CMFDA in two publications [Zhou et al. 2017; Shaw et al. 2018]. Shaw and colleagues stated, “Different imager vendors were evaluated and the Celigo imager was chosen for its high sensitivity in fluorescence imaging and clear, high-quality brightfield imaging.”
Learn more:
Single cell cloning (SCC) for cell line development

