Knowledge Collection
In order to select the appropriate cell counting method that is fit-for-purpose, researchers will need to first determine the intended use of the cell counting result, such as tumor digestion for single cell-based transcriptome analysis, mouse tissue processing for cytotoxicity assays, or isolation of human PBMCs for immunophenotyping analysis, etc. In addition, investigate to understand cell sample composition, where it is important to know the cell composition (various cell types, particle debris, chemical impurities, and suspension medium), as well as their visual appearances. The knowledge collected for the first two key factors can be used to hone in on the possible cell counting methods to select and evaluate.
Knowledge Collection
Determining the Intended Use of Cell Counting Results
Since the selection of cell counting methods should be fit-for-purpose, the first step is to understand or clarify the intended purpose of the measurements. While this may seem obvious at face value, it is important to think about the application of the cell counting method and what is ultimately to be gained. For example, a research lab may aim to document and publish novel biological processes or effects, and thus may seek to accumulate knowledge and incorporate best practices about the cell counting measurement quality early on and as a group. Meanwhile, cell therapy manufacturers may need to focus on meeting FDA requirements while being able to transfer assays from discovery to development, clinical trial sites, and manufacturing, ideally leveraging all assay improvements throughout the company. Both of these scenarios may also include multiple labs in multiple locations performing the same bioassays. Each of these goals require different considerations when it comes to choosing a method of cell counting.
Another critical component to determining intended use is what readout will be measured by the assay. This can be as simple as live or dead cell counts, but can also include measurements such as cell concentrations (alive or dead), percent viability, and biomarker or protein expression. These various metrics require different optimized conditions, especially when considered in the context of the sample type. The enormous range of biological sources for cell counting samples cannot be understated. From cultured or purified cell lines, whole tissues, isolated or cryopreserved blood samples, single-cell suspensions, to name a few, there are a plethora of considerations to choosing a cell counting method for each of these sample types. If manufacturing is the final goal, choosing the cell counting method will be dependent on the final product. For example, human bone marrow stromal cells for regenerative therapies have very different needs compared to tumor-infiltrating lymphocytes (TIL-T cells). Therefore, careful consideration of the intended use of the cell counting method must be employed to ensure the most robust and reliable results.
Researchers may download the Cell Counting Needs Assessment Worksheet or fill the worksheet on the website for determining the intended use of the cell counting results. Please contact Nexcelom if you would like to have one of our field application scientists review the assessment worksheet and provide guidance on selecting the best cell counting method for your needs.
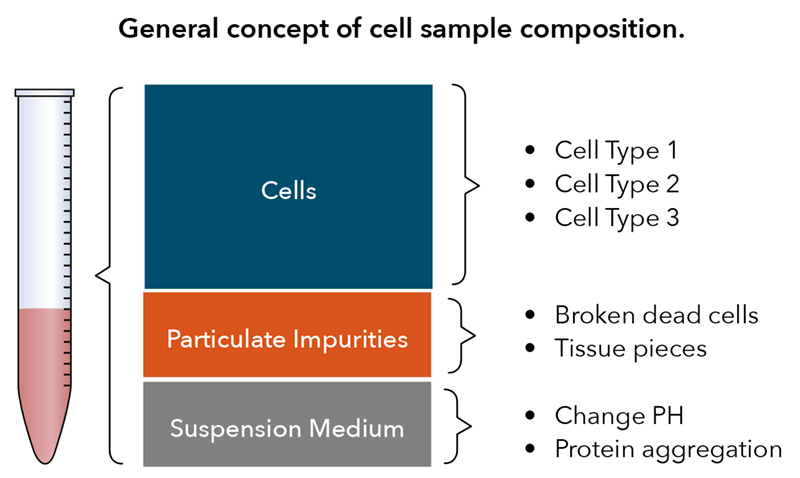
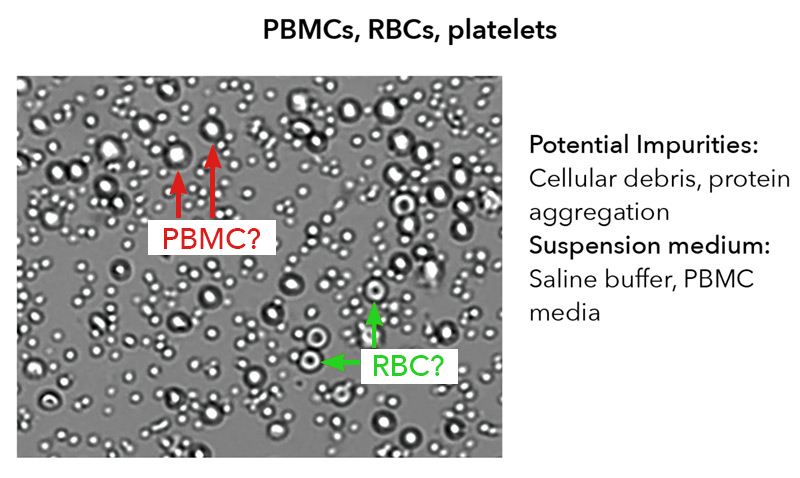
Figure 1. Cell sample composition. A fresh human PBMC sample is collected from a whole blood sample and processed by Ficoll separation and reveals: the cells of interest (PBMCs), but also platelets and RBCs, cellular debris, protein aggregates, and various types of culture media.
Understanding the Sample Composition
An important aspect of the knowledge collection process is to understand the composition of the cell samples. Perhaps most important is ascertaining what your cell samples visually look like, which is determined by what biological components are present. In general, a cell sample may contain various cell types, impurities such as debris or red blood cell (RBC) contamination, and the different suspension media used, all of which likely will affect cell counting procedures1. In the example provided, the cell sample contains three cell types, where only PBMCs are typically considered the measurands; thus, the cell counting method selected should be able to differentiate PBMCs from RBCs and platelets (Figure 1).
Cell morphologies can affect how well the cells are counted, where variation sizes, clumpiness of the cells can dictate the cell counting method to select (Figure 2). Additionally, cellular debris or protein aggregation and buffer/media selection may interfere with the staining process or cell counting, and therefore part of the method selection should ensure that these components will not interfere with the quality of cell counting. The examples provided in Figure 3 illustrate the types of debris or additional cell types that can be present in cell samples, aiding in the appropriate selection of a cell counting method to identify the intended measurands. Perhaps the most critical takeaway is that unpurified primary cell samples unquestionably require the use of fluorescent stains to identify the target cells of interest. The example images demonstrate the use of fluorescent nuclear staining using acridine orange and propidium iodide, which have been used as a live/dead dye mixture to fluorescently label only nucleated cells in green and red fluorescence respectively. Therefore, only nucleated cells will fluoresce and nonspecific particles such as debris, red blood cells, and platelets will not, thus correctly counting only the cells in the primary samples.
CHO S
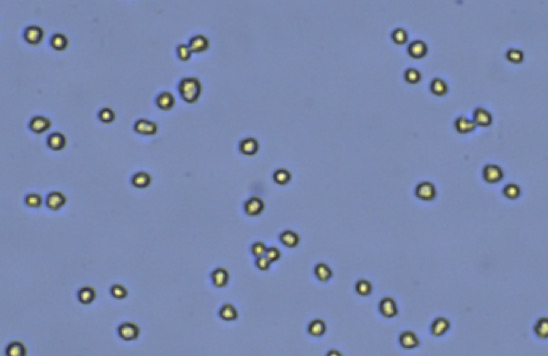
Uniform cell size and shape
UCAA-257
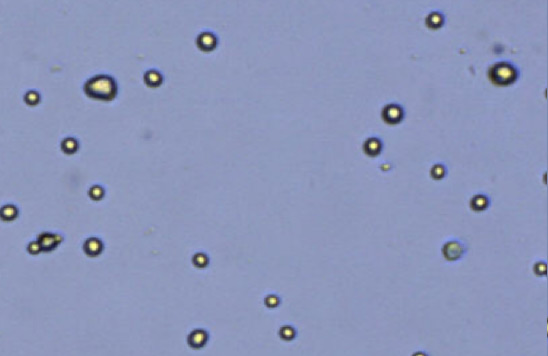
Heterogeneous cell sizes
CAKI
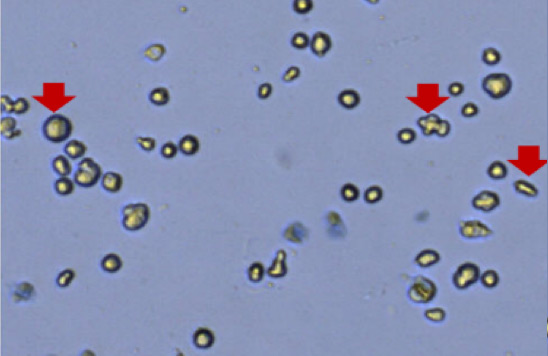
Large morphology variation
UCAA-257
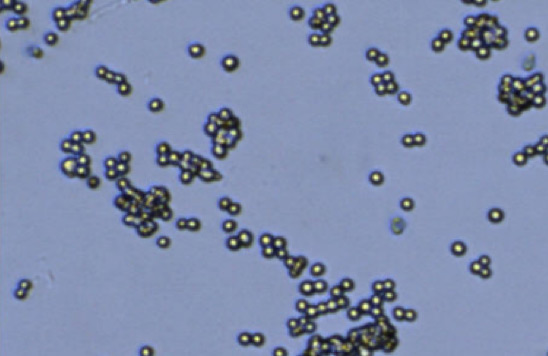
Clumpy
Figure 2. Commonly observed variation, debris, and additional cell types in samples.
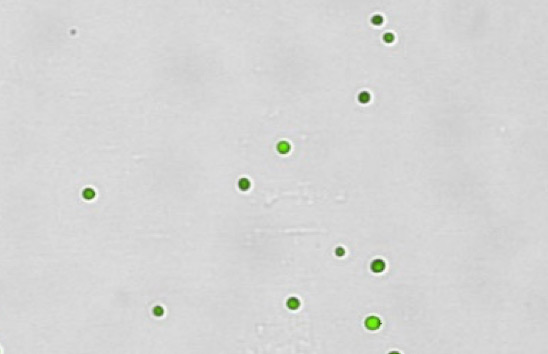
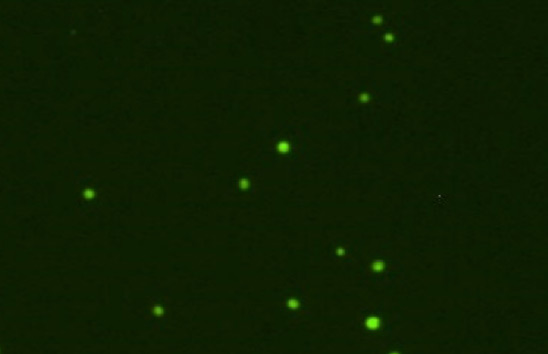
Purified CD34 cells
Clean, no debris, only nucleated cells
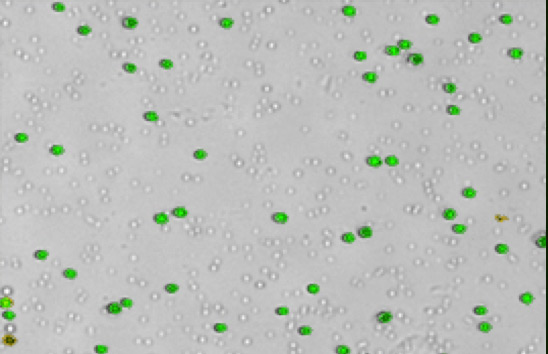
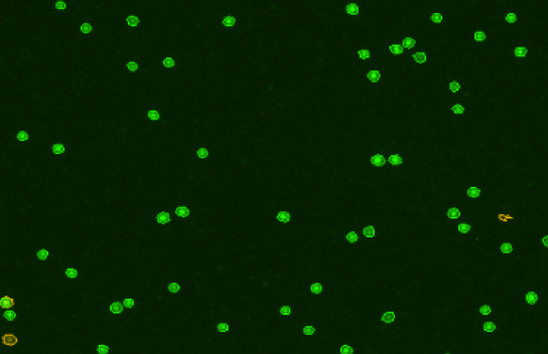
Ficolled human PBMCs
Some background debris, RBCs, platelets
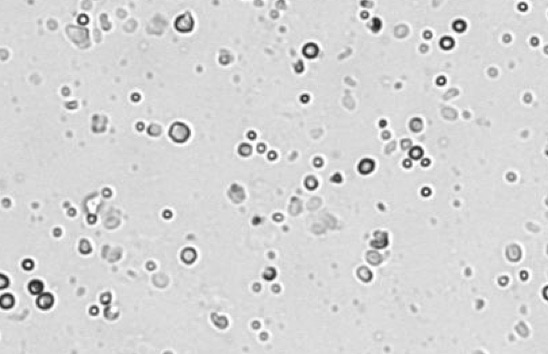
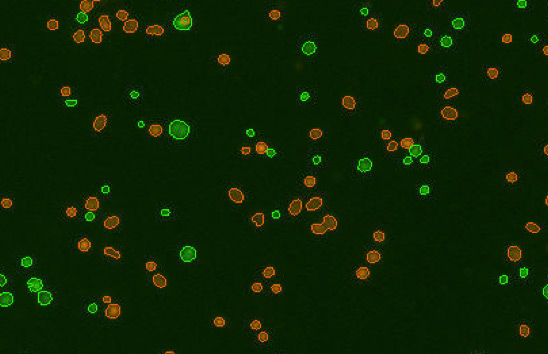
Human melanoma digest
Some background tissue debris, RBCs, platelets
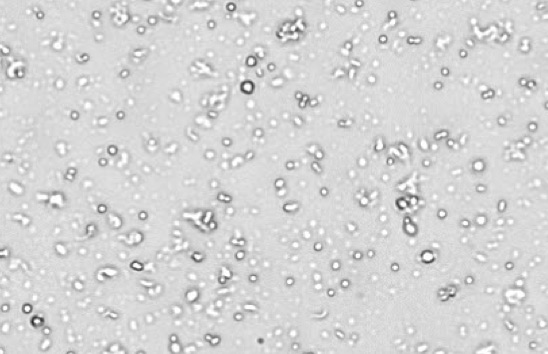
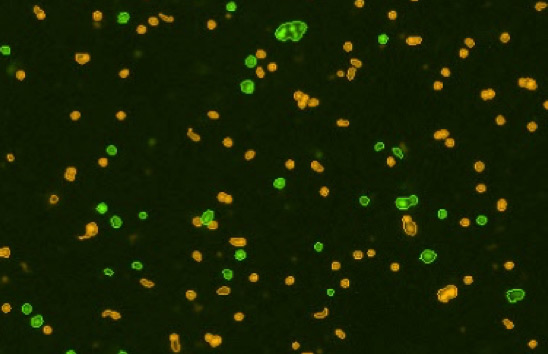
Primary lung digest
High background tissue debris, RBCs, platelets
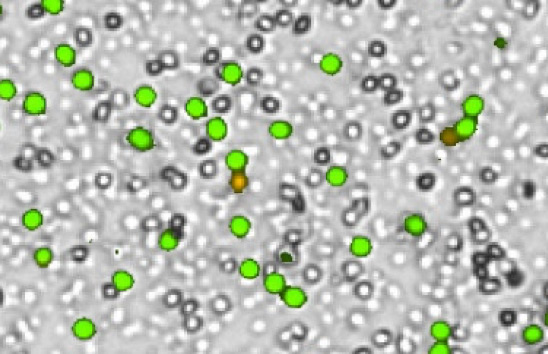
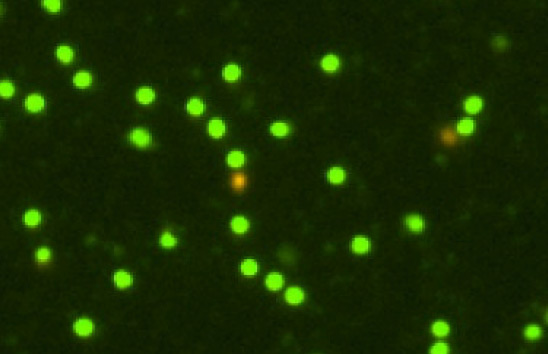
Primary human Leuko Pac
High background debris, RBCs, platelets
Figure 3. Commonly observed variation, debris, and additional cell types in samples.
References
- Chan LL, Laverty DJ, Smith T, et al. Accurate measurement of peripheral blood mononuclear cell concentration using image cytometry to eliminate RBC-induced counting error. J Immunol Methods. 2013;388(1-2):25-32.