High-throughput cell-based viral protein binding and inhibition assay
When developing vaccines or antiviral therapeutics it is important to identify the viral antigen or proteins responsible for the infection. Scientists will perform a viral protein binding and inhibition assay to screen a library of antibodies that can inhibit viral protein binding to the host cells. By allowing the target antibodies to interact with the viral antigen and host cells, the level of binding inhibition effects can be determined.
A high-throughput inhibition assay to study MERS-CoV antibody interactions using image cytometry
Citation https://www.ncbi.nlm.nih.gov/pubmed/30468747
There are currently 7 coronaviruses including the new emerging SARS-CoV-2 that are known to infect humans.
- HKU1-CoV
- 229E-CoV
- NL63-CoV
- OC43-CoV
- SARS-CoV
- MERS-CoV
- SARS-CoV-2
The emergence of Middle East respiratory syndrome coronavirus (MERS-CoV) poses serious threats to global public health and highlights the urgent need to rapidly identify and characterize potential neutralizing antibodies. MERS-CoV resulted in 2,494 confirmed infections and 858 deaths in 2019 globally at a high mortality rate of 35 – 40%.
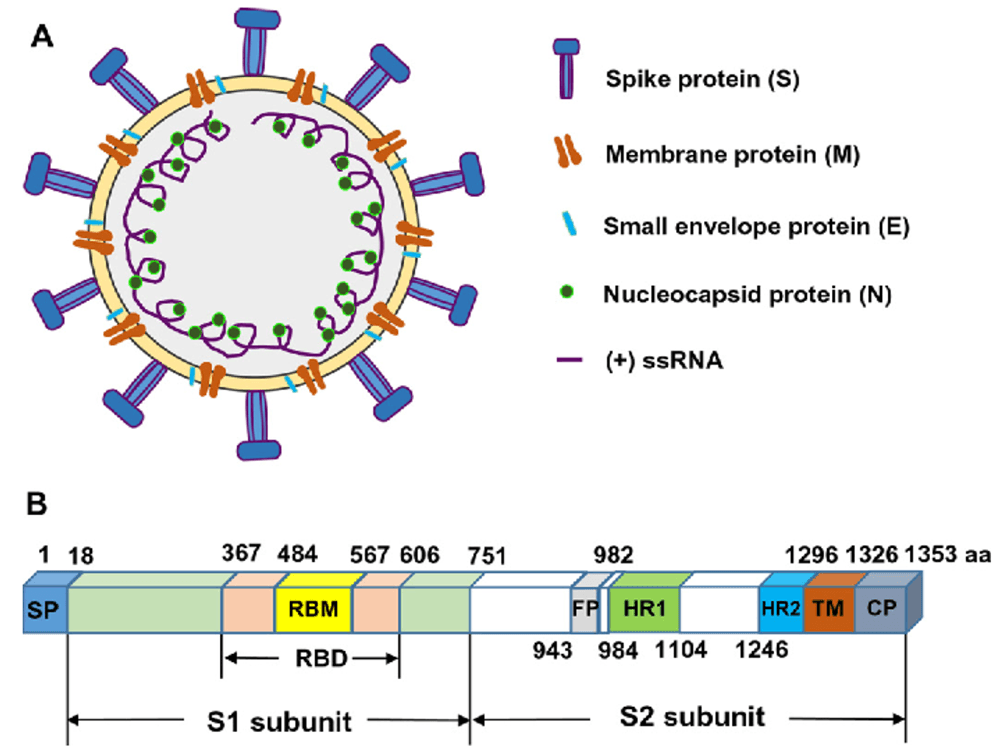
For MERS-CoV virions, the spike (S) proteins are present on the surface and mediate viral entry, thus making it the primary target for MERS-CoV vaccine and antibody development. The S proteins consist of S1 and S2 subunits, where the S1 region includes the RBD and NTD domains and can bind to the host cell receptor dipeptidyl peptidase-4 (DPP4). There is a need to identify monoclonal antibodies with broad neutralization activity towards RBD, NTD, and S2 binding sites.
Zhang, Naru et al. “Receptor-binding domain-based subunit vaccines against MERS-CoV.” Virus research 202 (2015): 151-9.
MERS-CoV display spike (S) proteins contain S1 and S2 subunits
Common methods for the detection of protein binding
Some of the commonly serological methods like ELISA, biolayer interferometry, and flow cytometry are used to study MERS-CoV antibody binding specificity and function.
Enzyme-Linked Immunoassay (ELISA)
ELISA is unable to assess antibody binding to antigen in their native conformation. In addition, unlimited access to epitopes may exaggerate antibody binding results, which can generate false-positive results.
Biolayer interferometry
Biolayer interferometry is limited by the requirement of tagged protein to immobilize to affinity tip. This detection method also may reduce sensitivity and reproducibility due to the difficulty in detecting bound proteins.
Flow cytometry
Flow cytometry can be time-consuming and tedious, which may not be practical for screening purposes. The system typically requires management by highly trained specialists, on-going maintenance by service engineers.
Cell-based antibody inhibition assay using the Celigo Image Cytometer
In this work, we present a high-throughput image cytometry method for protein binding inhibition assay. The high-throughput screening method used a cell type with high DPP4 expression, where optimal seeding density and protein binding conditions were determined in earlier experiments. The ability to detect the inhibition of MERS-CoV S binding to cells was tested across the titration of monoclonal antibodies. The binding inhibition results were comparable with previously published literature for MERS-CoV spike monomer and showed similar patterns for neutralization.
The Celigo Image Cytometer provides an efficient approach for characterizing potential therapeutic and prophylactic antibodies for combating MERS-CoV. The ability to rapidly determine direct antibody binding to host cells in a high-throughput manner can be applied to study other pathogen-antibody interactions and thus can impact future research on viral pathogens.
Celigo images showing the fluorescent image (left) and the counted image (right).
The Celigo is used to evaluate binding inhibition of 3 antibodies, D12, G2, and G4, which recognize MERS-CoV RBD, NTD, and S2, respectively. The assay is optimized by selecting the appropriate cell type, optimal seeding density, and the suitable MERS-CoV viral protein.
BHK21 is selected for optimized cell type expressing DPP4
The cell types A549, BHK21, and VeroE6 are transfected with DPP4 expression. Each cell type is transfected with 0.02 to 2 µg of DPP4 plasmid. The transfected cells are then labeled with DPP4 PE-conjugated mouse antibody (red fluorescence). The BHK21 cell line demonstrated the highest DPP4 expression with the brightest fluorescence from imaging and fluorescent histogram.
Red fluorescent images showing DPP4 expression for 3 cell types, as well as the fluorescent intensity histogram showing bright signals for BHK21
Seeding density of 5 x 103 cells/well is selected
Three different seeding densities are tested at 2 x 104, 5 x 103, and 1 x 103 cells/well. The 2 x 104 cells/well is too crowded and 1 x 103 is too few for optimized assay.
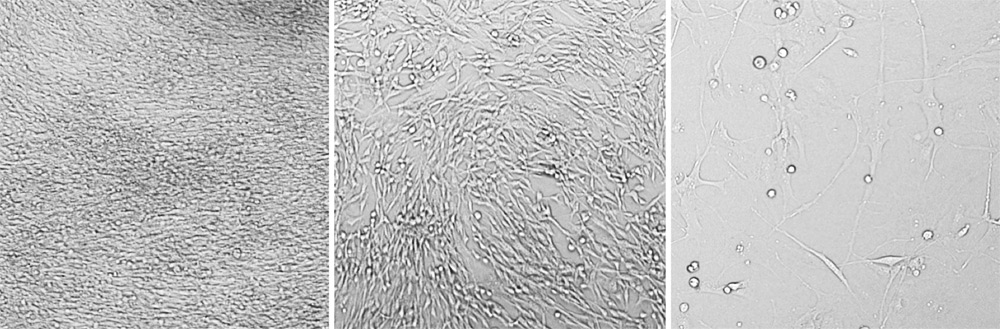
Bright field images showing the confluence of host cells in the wells. Show from left, 2 x 104 cells/well, 5 x 103 cells/well, 1 x 103 cells/well
Selected MERS-CoV S trimer for the antibody binding inhibition assay
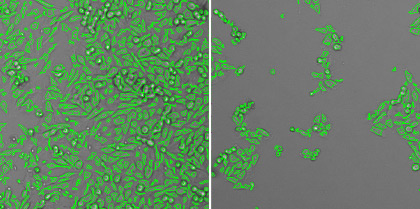
Two MERS-CoV viral antigens are tested, MERS-CoV S1 trimer and MERS-CoV S monomer proteins. The proteins are titrated from 0.1 to 10 µg. The viral proteins are allowed to bind to the host BHK21 cells and then labeled with Alexa Fluor 488-MERS-CoV Spike antibody. The 2 µg produced the highest signal-to-background ratio.
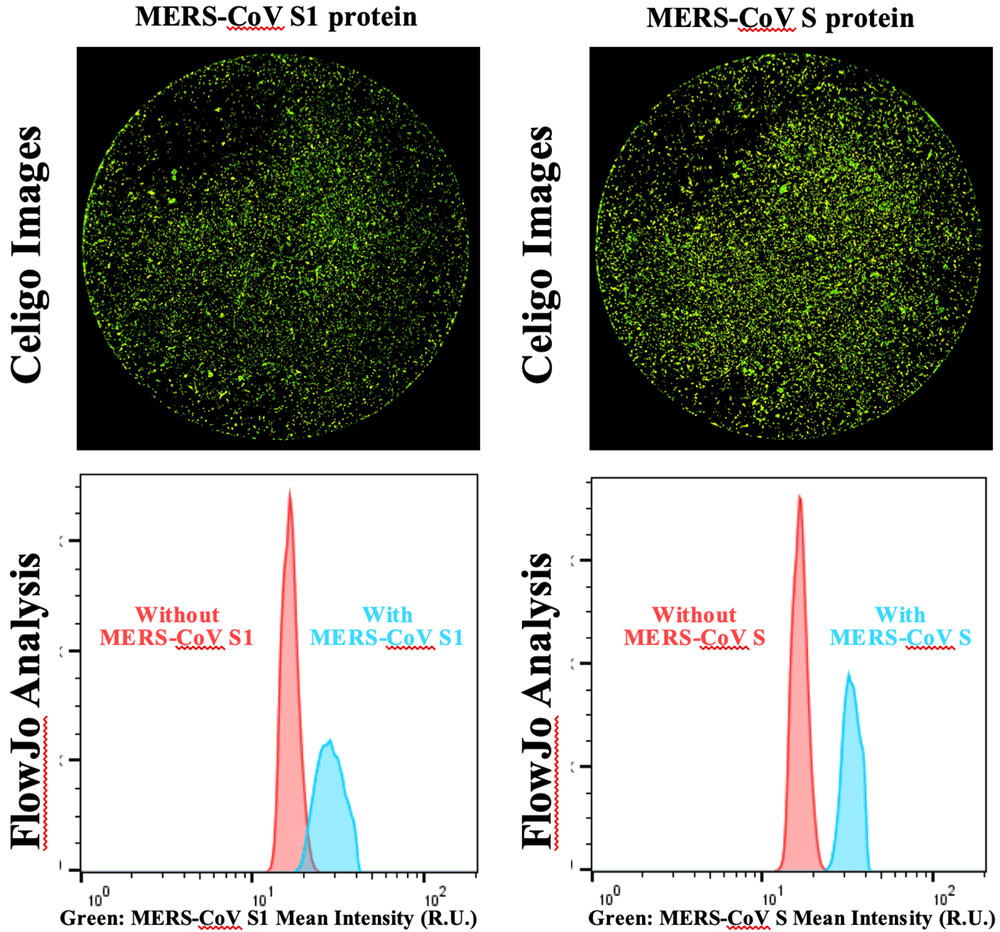
Green fluorescent images for MERS-CoV S1 and MERS-CoV S protein binding to BHK21 host cells, as well as fluorescent intensity histograms with MERS-CoV S showing the largest signal-to-background separation.
Cell-based antibody binding inhibition assay
In this experiment, BHK21 cells are seeding into a 96-well plate at 5 x 103 cells/well and allowed to incubate overnight. 2 µg/well of MERS-CoV S protein is incubated with 4-fold serial dilutions of 200 µg/mL of G2, D12, or G4 monoclonal antibody (mab) for 30 min at room temperature. The mixture of the MERS-CoV S and mab is then added to the DPP4-expressing BHK21 cells and incubated for 2 hours at room temperature. Finally, the cells are washed and fixed with cold acetone, and then stained with AF488-goat-anti-rabbit IgG and DPP4 PE mouse ab, as well as 300 nM of DAPI.
The Celigo is used to acquire the bright field and fluorescent images to analyze MERS-CoV binding directly to the BHK21 host cells. The DAPI signal is used to identify cells and analyze AF488 fluorescence.
Fluorescent overlay images of the DAPI and AF488 fluorescence for D12 antibody dose-dependent inhibition results. As D12 concentration increased, green fluorescent signal decreased.
Both D12-RBD specific mab and G2-NTD specific mab showed dose-response inhibition effects to the binding of MERS-CoV S protein on to the host cells. The G4-S2 specific mab did not show any inhibition to the binding.
Dose-response curves for D12, G2, and G4 mabs.
Luciferase-based viral neutralization assay confirming the results for D12 and G2 mabs
A luciferase-based neutralization assay is conducted to confirm the image-based antibody binding inhibition results. The D12 and G2 mab interfered with the binding of MERS-CoV S protein to DPP4, demonstrating neutralization effects of the virus. The G4 also showed a neutralization effect at higher IC50 values that did not correlate to the binding assay. This may be due to that G4 neutralizes by interfering with the fusion of MERS virus into the host cells instead of direct binding.
Learn more about modern virology assays: