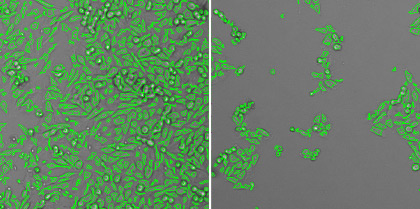
Direct counting of individual infected cells labeled with mKate2-expressing influenza virus
Researchers working with replication-incompetent viruses will require the use of fluorescent viral particles and fluorescent immunostaining to detect single infected cells. In order to quantify, the flow cytometry method is typically used to analyze 1000 – 2000 host cells in 96- or 384-well plates. First, these host cells require the lengthy process trypsinization which disturbs the host and virus interaction. Next, flow cytometry requires approximately 2 minutes per sample to analyze the infected cells, which is equivalent to 3 hours for 96 samples and around 12 hours for 384 samples.
The Celigo Image Cytometer can efficiently image and quantify individual infected cells, where the system can scan and analyze an entire 384-well plate in less than 10 min/plate in bright field (HRP labeling) and fluorescence. The images below show individual infected MDCK cells with mKate2-expressing influenza virus. The plate imager was able to directly count the infected cells within a single well without trypsinization, calculate infection rate, and generate a highly linear viral titration plot using the equation
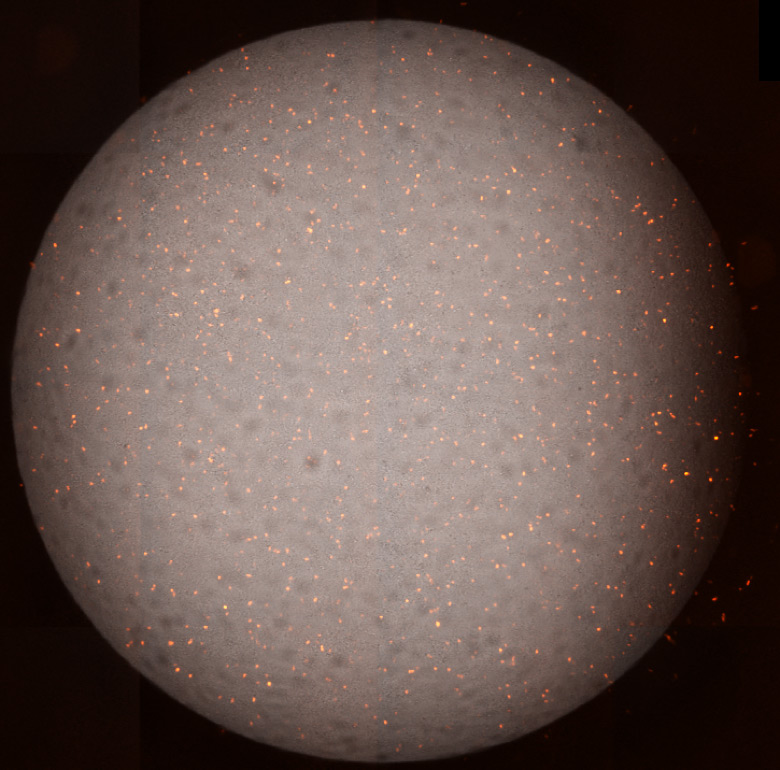

Whole well bright field and fluorescent overlay images for high (left) and low (right) viral concentrations
| Dilution | 125 | 625 | 3125 | 15625 |
| mKate2+ counts | 1530 | 333 | 63 | 11 |
| mKate2 % | 2.29% | 0.43% | 0.09% | 0.02% |
Direct counting results of mKate2 positive infected MDCK cells, where the % infection was calculated from the total cell count
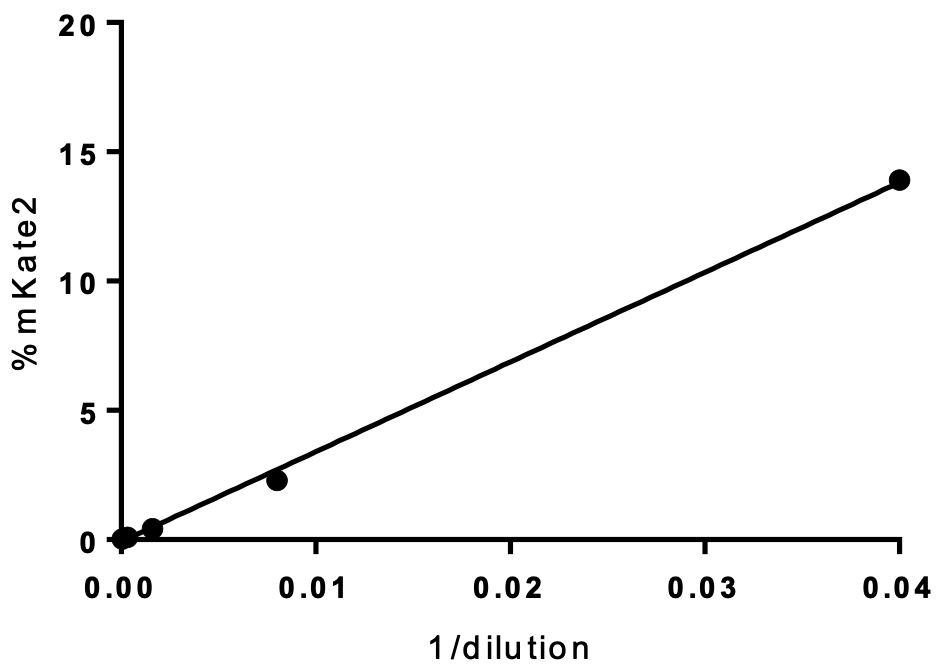
Influenza viral infection rate in respect to dilution
The Celigo Image Cytometer automates viral titer assays:
Direct counting of individual infected cells
Learn more about modern virology assays: