A rapid method to establish potential molecular targets for colorectal cancer progression
Colorectal cancer (CRC) is the second leading cause of cancer mortality and accounts for nearly 8% of all new cancer cases in the United States [1]. The total number of cases is steadily falling by an average of 2.0% year over year owing, in large part, to advances in screening awareness, diagnostics, and therapeutics. However, tumor heterogeneity can lead to complications from traditional chemotherapy and promote drug resistance [2]. The development of targeted molecular therapies is an increasingly popular approach to combat and overcome the limitations of conventional treatments.
Investigations into the underlying molecular mechanisms responsible for the development and progression of CRC may elucidate new therapeutic targets to mitigate the downsides of conventional treatments such as drug resistance. Tong et. al. recently performed a series of illuminating experiments to identify the function and molecular mechanisms of dipeptidyl peptidase 3 (DPP3) in CRC. A combination of clinical tissue samples, in vitro human CRC cell lines, and in vivo target gene knockdown models were all investigated in this study.
High-throughput Measurements and Characterization
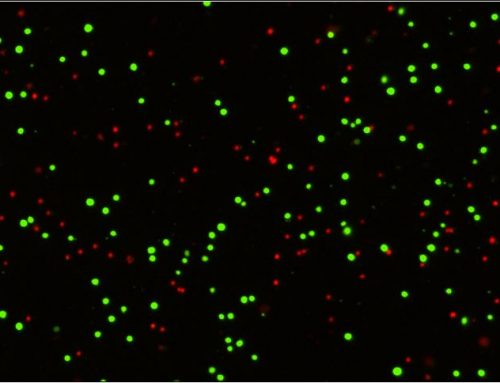
Bioluminescence imaging and quantification of the tumor burden were measured using the IVIS® Spectrum in vivo imaging system (Revvity). Immunohistochemical staining of paraffin-embedded human CRC biopsies provided further evidence supporting an active role of DPP3 in the progression of CRC and overall patient prognosis. Downregulation of DPP3 via lentiviral expression of DPP3-targeted shRNA in CRC cell lines was characterized by measuring cell proliferation (Celigo Image Cytometer, Nexcelom Bioscience), apoptosis, and migration. RNA sequencing was also used to explore the molecular mechanism of DPP3 responsible for promoting the progression of CRC. Collectively, these experiments helped to establish the role of DPP3 in both the development and progression of CRC by targeting CDK1.
Rapidly Image Multiple Cell Lines Using Various Cell-Based Assays
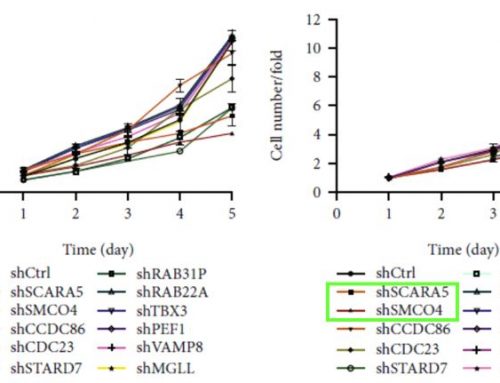
Multiple human CRC cell lines, primarily HCT 116 and RKO, were used to investigate the role of DPP3 and CDK1 in the development and progression of CRC. Target gene knockdown models were designed using human DPP3 and CDK1 genes as templates and knockdown efficiencies were verified by qPCR and western blots. The HCT 116 cells with the depletion of CDK1 (shCDK1) and the combined knockdown of DPP3 and CDK1 (shCDK1 + shDPP3) examined the combined effects on CRC. The Celigo system counted the individual cells in each well daily over five days to measure the proliferation rate for each experimental condition (Figure 1). Depletion of CDK1 significantly reduced cell proliferation and the combined depletion of DPP3 and CDK1 further inhibited growth in the CRC model. These findings were also supported with additional cell-based assays to measure colony formation, apoptosis, cell migration, and wound-healing using complementary experimental techniques.
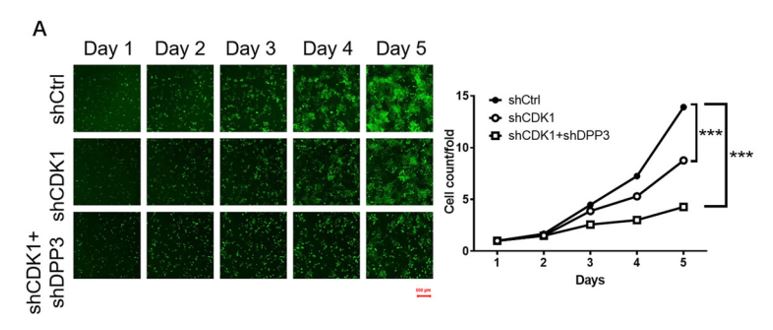
Figure 1. Knockdown of DPP3 deepens the effects on CRC cells by CDK1 knockdown. Cell models were subjected to the detection of cell proliferation by a Celigo cell counting assay. (Modified from Tong et. al. Cell Death and Disease (2021))
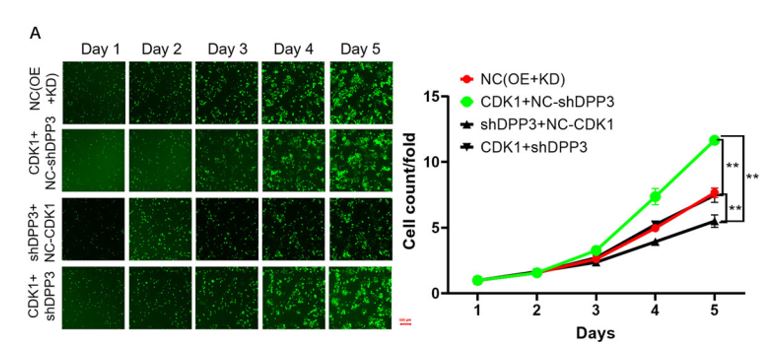
Figure 2. Overexpression of CDK1 alleviates the inhibitory effects of DPP3 knockdown in CRC cells. Cell models were subjected to the detection of cell proliferation. (Modified from Tong et. al. Cell Death and Disease (2021))
High-Throughput In vivo Imaging
RKO cells with or without the DPP3 knockdown were injected into mice to perform functional validation of DPP3 in vivo with 10 animals in each group. Collection of the tumor burden data occurred 2-3 times per week following the tumor inoculation for up to 25 days. Bioluminescence imaging (IVIS® Spectrum system, Revvity) established a reduction in the overall tumor burden in the shDPP3 group and a reduction in the bioluminescence intensity (Figure 3). Ki67 staining of excised tumors from the mouse model exhibited reduced cell proliferation in the shDPP3 samples indicating the downregulation of DPP3 inhibits CRC tumor growth in vivo.
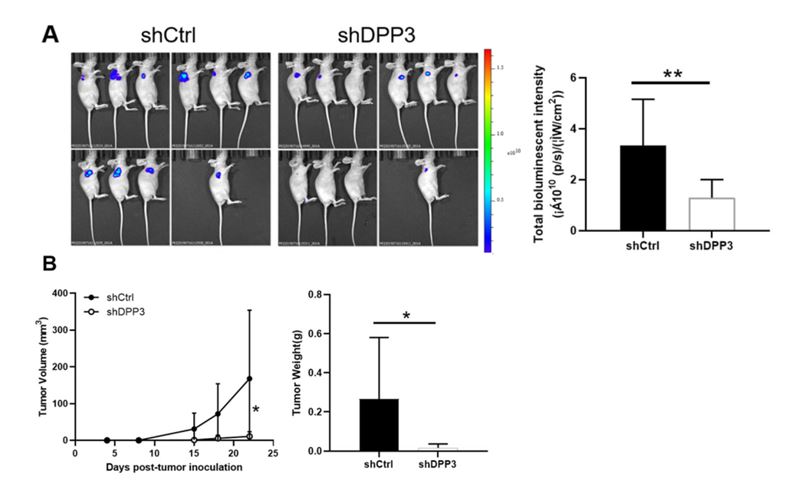
Figure 3. DPP3 knockdown inhibited CRC development in vivo. (A) In vivo imaging was performed to evaluate the tumor burden in mice of shDPP3 and shCtrl groups post-tumor-inoculation; (B) The bioluminescence intensity was scanned and used as a representation of tumor burden in mice of shDPP3 and shCtrl groups. (Modified from Tong et. al. Cell Death and Disease (2021))
Immunohistochemistry (IHC)
IHC was performed on patient biopsy samples (99 CRC samples, 76 normal samples) to characterize the role of DPP3 expression in CRC. Heightened expression levels of DPP3 in tissue samples predicted poor prognosis for CRC patients (Figure 4). A positive correlation with clinical indications such as lymphatic metastasis, pathological stage, and a positive number of lymph nodes was observed with elevated DPP3 expression. Finally, Kaplan-Meier survival analysis indicated that high expression of DPP3 in tumor tissue predicted a reduction in overall survival time.
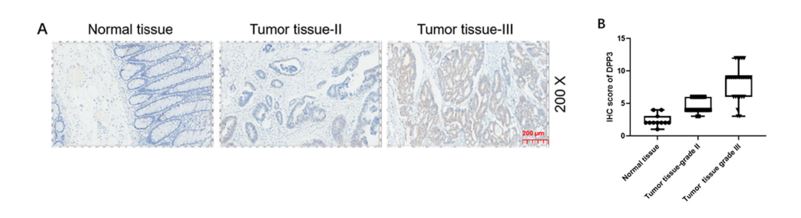
Figure 4. DPP3 was upregulated in CRC and knockdown of DPP3 cells models was constructed. A, B The expression level of DPP3 was detected by IHC analysis in normal tissues and CRC tissues with different T stages. (Modified from Tong et. al. Cell Death and Disease (2021))
The combination of in vitro experiments, in vivo tumor characterization, and analysis of clinical tissue biopsies from human patients provides strong support for the involvement of DPP3 in the development and progression of CRC by targeting CDK1. The experiment also provided evidence for the synergistic effects of simultaneous depletion of DPP3 and CDK1 resulting in greater inhibition of CRC development and growth.
The imaging and quantification of the CRC cell culture models on the Celigo Image Cytometer, along with the characterization of the target genes for DPP3, provided a rapid method to establish potential molecular targets for further investigation in vivo and inpatient biopsy samples. It is worth noting that the Celigo is also capable of performing the supporting cell-based assays that were demonstrated in this study (apoptosis, migration, wound-healing, and cell cycle analysis). The identification of molecular targets is a critical stage in developing effective new therapeutics or high-throughput screening of candidate molecules. These stages of development are routinely performed on the Celigo platform to leverage the rapid screening capabilities and integrated software analysis tools all in a convenient benchtop instrument. The optional 21 CFR Part 11 software also facilitates compliance with regulatory documentation requirements.
The Celigo Image Cytometer offers a wide variety of applications for automated cell counting and cell-based assays with intuitive software analysis tools and automation options to accommodate high-throughput needs. Learn more about cellular models and the direct cell counting assays on our Celigo Image Cytometer Applications page or contact us directly to book a seminar or live demonstration today.
References
- Siegel, R. L., Miller, K. D., Goding Sauer, A., Fedewa, S. A., Butterly, L. F., Anderson, J. C., Cercek, A., Smith, R. A. & Jemal, A. Colorectal cancer statistics. CA Cancer J Clin 70, 145–164. https://doi.org/10.3322/caac.21601 (2020)
- Punt, C. J., Koopman, M. & Vermeulen, L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Rev. Clin. Oncol. 14, 235–46. (2017).
- Tong, Yixin et al. “DPP3/CDK1 contributes to the progression of colorectal cancer through regulating cell proliferation, cell apoptosis, and cell migration.” Cell death & disease vol. 12,6 529. 22 May. 2021, doi:10.1038/s41419-021-03796-4


Leave A Comment