Introduction
Chinese hamster ovary (CHO) cells are the cell type of choice for biopharmaceuticals due to their propensity to correctly fold and post-translationally modify human proteins [1]. Here, researchers use an optimized bioinformatics tool (“CRISPy”) to generate chimeric RNA called “single guide” RNA (sgRNA) in order to disrupt FUT8, a gene that is utilized in N-glycosylation. This novel web-based tool ensures efficient, fast, and low-cost genetic manipulation of CHO cells for future biopharmaceutical applications.
Materials and Methods
Cell culture and transfection of CHO cells
CHO cells were cultured and transfected by Nucleofector Kit V with 1 μg of Cas9 plasmid and 1 μg of sgRNA plasmid. Two days later, cells were transfected with pmaxGFP and assessed for transfection efficiency using Nexcelom’s Celigo.
Phenotypic analysis of FUT8 knockout cells
Five days post-transfection, the selection agent Lens culinaris agglutinin (LCA) was added. (LCA binds plasma membrane proteins, which leads to downstream cell death. LCA serves as a selection agent for FUT8 knockout cells as LCA cannot bind to cells devoid of FUT8.) Cells were stained for phenotypic analysis with Hoechst (red in image) and fluorescein-labeled LCA (F-LCA; green in image). FUT8 wild-type cells will stain green, while FUT8 knockout cells will not. Bright-field and fluorescent images were captured with the Celigo.
*Note: For more detailed Materials and Methods and a complete account of the entire study, please refer to the original manuscript [http://onlinelibrary.wiley.com/doi/10.1002/bit.25233/abstract]
Results
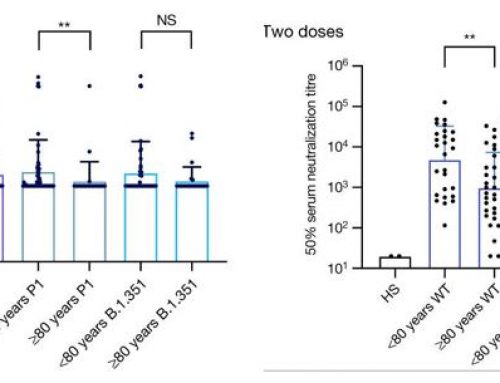
- LCA selection caused control cells to become non-adherent and rounded.
- Cells transfected with Cas9 and sgRNA remained adherent, indicating the successful knockout of FUT8 in those cells.
- Cells transfected with Cas9 and sgRNA without LCA selection showed that F-LCA negative cells represented 29.1% of the entire population.
- Cells transfected with Cas9 and sgRNA with LCA selection showed 98.6% of cells were F-LCA negative, as demonstrated by other studies proving the lack of functional FUT8 [2-4].
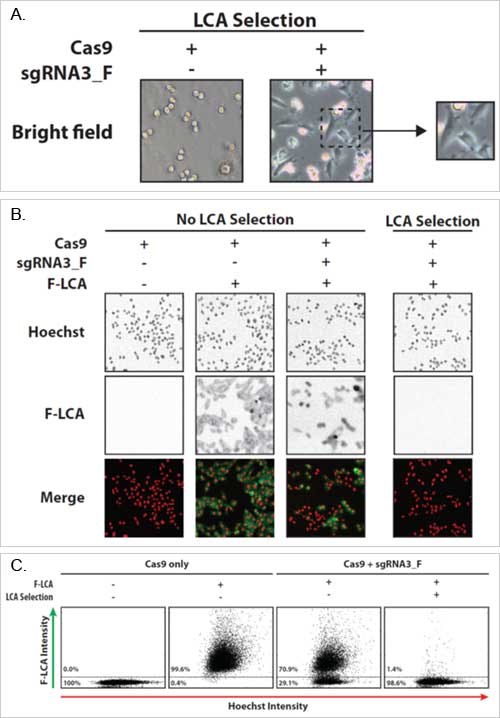
Figure 1A-C. Phenotypic analysis of FUT8 knockout cells. Cells were transfected with either Cas9 plasmid alone or Cas9 plasmid plus sgRNA. At Day 6 the bright field images were taken (A). On Day 13 the cells were stained with Hoechst (red) and fluorescein-labeled LCA (green). Quantification of the images for F-LCA positive (wild-type FUT8) and F-LCA negative (FUT8 knockout) can be seen in C.
Conclusions
- This RNA-guided CRISPR Cas9 application successfully disrupted genes in CHO cells.
- The web-based bioinformatics design tool “CRISPy” provides an effective, low-cost method for genetically manipulating CHO cells.
References
- Jayapal, K.P., et al., Recombinant protein therapeutics from CHO cells – 20 years and counting. Chem Eng Prog, 2007. 103: p. 40-47.
- Malphettes, L., et al., Highly efficient deletion of FUT8 in CHO cell lines using zinc-finger nucleases yields cells that produce completely nonfucosylated antibodies. Biotechnol Bioeng, 2010. 106(5): p. 774-83.
- Mori, K., et al., Engineering Chinese hamster ovary cells to maximize effector function of produced antibodies using FUT8 siRNA. Biotechnol Bioeng, 2004. 88(7): p. 901-8.
- Yamane-Ohnuki, N., et al., Establishment of FUT8 knockout Chinese hamster ovary cells: an ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol Bioeng, 2004. 87(5): p. 614-22.

Leave A Comment