Transcript: Assay Show: Measure Apoptosis Using Caspase 3/7 for Adherent MDA-MB-231 Cells Using Celigo Image Cytometer
Welcome to the assay show. This video will show you how to perform an apoptosis assay on the Celigo image cytometer using caspase 3/7 and Hoechst reagents
Let me first take a moment and describe the assay principle.
- DEVD is a caspase 3/7-specific sequence that is coupled with a DNA dye molecule.
- This substrate can freely diffuse across the cell membrane in live cells.
- Once inside apoptotic cells, the caspase 3/7 protein recognizes and cleaves the DEVD sequence and releases the DNA probe.
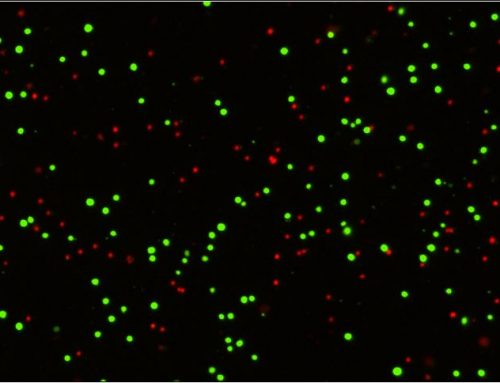
- When the probe enters the nucleus it binds to the DNA producing a bright green fluorescent signal.
- Hoechst dye was added as a nuclear counterstain for the detection of total nucleated cells
After staining, the Celigo was used to acquire whole well bright field, Caspase 3/7-Green, and Hoechst -Blue images.
The Celigo software automatically analyzes the captured images and reports the total number of green, caspase positive cells the total number of blue, Hoechst positive nucleated cells and the percent of apoptotic cells. The captured bright field images were not analyzed and were used to monitor cell morphology.
Today I will show you an apoptosis assay that was performed using an adherent breast cancer cell line MDA-MB-231 that was treated with 3 micromolar staurosporine.
To achieve the best accuracy of your cell plating, measure the cell concentration and viability using a Cellometer automated cell counter.
Mix 20 microliters of cell sample and 20 microliters of trypan blue
Load 20 microliters of stained sample into the cellometer chamber slide and perform a cell count to acquire cell number, concentration and viability.
Based on the measured concentration of your cells, per well, plate 10,000 cells in a volume of 200 microliters and allow the cells to incubate overnight
Next day add 3 micromolar staurosporine to induce apoptosis and allow the cells to incubate for the next 8 hours.
After the incubation, remove 100 microliters from each well.
And add 100 microliters of 2x concentrated Caspase 3/7 and Hoechst staining solution to each well
Allow the plate to incubate at 37 degrees for 30 minutes.
Because the Celigo can image and analyze adherent cells, trypsinization is not required.
Whole-well imaging of 24 wells was captured in three and a half minutes.
The analyzed results are displayed in a plate-based format showing a thumbnail picture and percent of apoptotic cells for each analyzed well.
Let’s take a closer look at a treated sample in well D3. By double clicking on a well the whole well image appears for review.
We can zoom in to look at the cell morphology in the bright field image, examine the staining and counting of caspase and Hoechst positive cells. In this merged image, the blue outlines represent the Hoechst positive total cells, and the red outlines represent the caspase 3/7 positive cells.
Celigo built-in gating interface provides the necessary tools for plotting and analyzing 2D scatter plots. Optimized setting are then automatically applied to the entire plate for easy data analysis.
All the data can be exported to excel as a .CSV file in a plate-based layout.
Each excel file contains the number of caspase 3/7 and Hoechst positive cells as well as a percent of apoptosis.
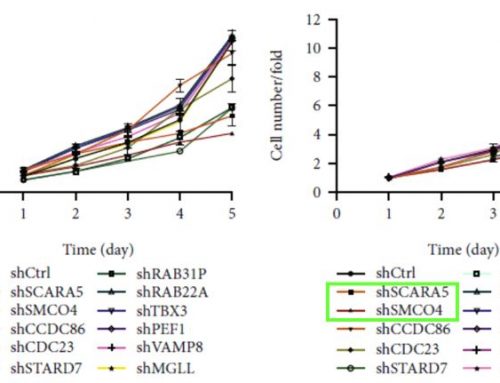
Generated bar graphs show an increased number of caspase 3/7 positive cells and percent apoptosis in the staurosporine-treated samples.
These and other assays are routinely performed on the Celigo. To learn more or schedule a free in-lab demonstration call us or visit nexcelom.com


Leave A Comment