Infectious viral outbreaks such as SARS, MERS, H1N1, Zika, and the current ongoing severe acute respiratory syndrome-related Coronavirus COVID-19 have significantly impacted the health and safety of communities around the world. The continuing outbreak of the SARS-CoV-2 virus has sparked the global research community to rapidly develop potential vaccines and antiviral therapeutics to aid in the prevention, reduce viral replication, or enhance immune response.
Types of Coronaviruses
Coronaviruses (CoVs) get their name from the Latin word “corona” which means “crown”, after the Spike protein (S) found on the surface of the virus. There are four common coronaviruses that commonly circulate in humans and are responsible for cold-like symptoms: HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1. Three other CoVs have emerged as infectious agents, jumping from their normal animal host species to humans: SARS-CoV, MERS-CoV and most recently, SARS-CoV-2 (1).
Why are Leading Virology Labs Working with Nexcelom to Drive the Development of Modern Viral Assays?
Development of Modern Virology Assays
In the past few years, Nexcelom Bioscience has worked with leading labs in the field of virology from government agencies, academic labs, and pharmaceutical companies such as the Centers for Disease Control and Prevention (CDC), Food and Drug Administration (FDA), and National Institute of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID). These labs have developed modern virology assays utilizing plate imagers to increase sensitivity, efficiency, and accuracy which has led to a reduction of materials used and reduction in time to answer critical questions.
Our dedicated field application scientists provide support for the development of new and novel high-throughput image cytometry assays for vaccines and antiviral therapeutics investigation.
How the Celigo Image Cytometer Can Speed Up Vaccine Development in Your Lab
Image Cytometry for Vaccine Development
The Celigo Image Cytometer can fast track vaccine development process by conducting assays in 96- or 384-well plates based on enumeration of foci, plaque or single infected cells. The Celigo provides the best-in-class whole well imaging in bright field and 4-color fluorescence, where the foci, plaque or single infected cells can be segmented and counted as images are taken and data is quickly available for you to make your next decision.
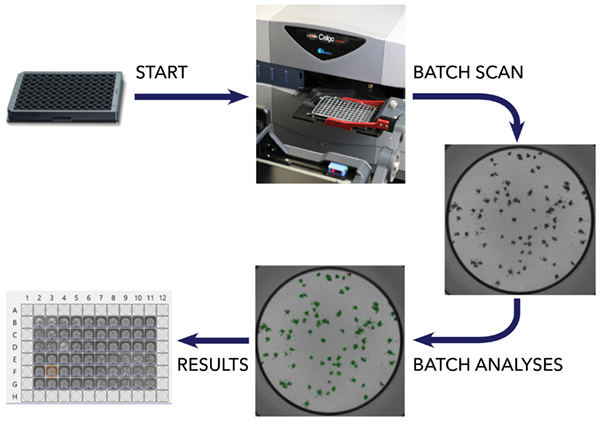
Automation of Viral Infectivity Assays at the Single Infected Cell Level
The ability to increase throughput by automating testing offers many benefits, including:
- Increased Productivity: Rapidly analyze data from viral titer and micro-neutralization assays— 50 plates (96- or 384-wells) in a single day
- Reduced Costs: Optimize your assay to reduce start material volumes in a 384 micro-titer plates
- Improved Performance: Increased sensitivity with detection of single infected cells
- Time Savings: Reduce assay duration with early detection of viral infection
- Flexibility: Co-cultured host cells viral infectivity assessment with the absence or presence of viral receptor
| Plate Type | Images/Wells | Typical Time (min) |
|---|---|---|
| 1536-well | 1 | < 6 min |
| 384-well | 1 | < 2 min |
| 384-well | 4 | < 5 min |
| 96-well | 16 | < 3.5 min |
Streamline Your Virology Assays
- Power in Resolution: Gain Single cell level sensitivity by image acquisition 1 um/pixel
- Flexible to Your Workflow Needs: Tailored acquisition and analysis software suite for your virology assays involving viral plaques, foci, and single infected cells in brightfield and fluorescence
- Multiple Readouts from a Single Plate: Morphological and cell health quantification to automate cytopathic effect (CPE) analysis
A Single Tool to Complete Several Critical Workflows in Viral Pathogen Research
Below are some examples of how the Celigo has been used in modern virology labs:
- Screening for monoclonal antibodies to block virus entry into host cells
- Screening for small molecules to disrupt viral replication inside host cells
- Investigating the interaction of virus and host cells
- Testing serological samples for viral titer and neutralization
- Investigating host immune responses
What High-throughput Virology Assays Can Be Run on the Celigo?
The Celigo Image Cytometer reliably enables high-throughput assays that aid in data driven decisions with confidence for your next move in the lab. Assays that can be run on the Celigo include:
- Viral Titer Assays
- Neutralization Assays
- Binding and Inhibition Assays
- Infectivity and Screening Assays
- Immune Monitoring and Response Assays
- CPE Assays
- Compound Cytotoxicity Assays
- Host Virus Interaction Assays
1. Centers for Disease Control and Prevention, U.S. (2020) Coronavirus: About Human Coronavirus Types. [Internet: https://www.cdc.gov/coronavirus/types.html Accessed February 10, 2020]
Publications using the Celigo Image Cytometer
Rosen et al. (2019) “A high-throughput inhibition assay to study MERS-CoV antibody interactions using image cytometry“, J Virol Methods, 265:77-83
Li et al. (2013) “A short hairpin RNA screen of interferon-stimulated genes identifies a novel negative regulator of the cellular antiviral response“, mBio, 265:77-83
Behzadi et al. (2019) “An Influenza Virus Hemagglutinin-Based Vaccine Platform Enables the Generation of Epitope Specific Human Cytomegalovirus Antibodies“, Vaccines (Basel), 7(2)
Bailey et al. (2019) “Antibodies Elicited by an NS1-Based Vaccine Protect Mice against Zika Virus“, mBio, 10(2)
Taverner et al. (2019) “Calcium Influx Caused by ER Stress Inducers Enhances Oncolytic Adenovirus Enadenotucirev Replication and Killing through PKCα Activation“, Mol Ther Oncolytics, 15:117-130
Stein et al. (2019) “CD46 facilitates entry and dissemination of human cytomegalovirus“, Nat Commun, 10(1):2699
Martinez et al. (2018) “Characterizing the Different Effects of Zika Virus Infection in Placenta and Microglia Cells“, Viruses, 10(11)
Charman et al. (2019) “Constitutive TRIM 22 expression within the respiratory tract identifies 2 tissue-specific and cell-type dependent intrinsic immune barriers to 3 influenza A virus infection 4“, bioRxiv
Qin et al. (2019) “Effect of Periostin Silencing on the Autophagy of Osteoblasts“, Cell Reprogram, 21(3):122-128
Yang et al. (2017) “IL-6 ameliorates acute lung injury in influenza virus infection“, Sci Rep, 7:43829
Ramos et al. (2019) “Innate Immune Response to Influenza Virus at Single-Cell Resolution in Human Epithelial Cells Revealed Paracrine Induction of Interferon Lambda 1“, J Virol, 93(20)
Masci et al. (2019) “Integration of Fluorescence Detection and Image-Based Automated Counting Increases Speed, Sensitivity, and Robustness of Plaque Assays“, Molecular Therapy Methods & Clinical Development, 14:270-274
Panda et al. (2019) “IRF1 Maintains Optimal Constitutive Expression of Antiviral Genes and Regulates the Early Antiviral Response“, Front Immunol, 10:1019
Baldwin et al. (2018) “Purified Inactivated Zika Vaccine Candidates Afford Protection against Lethal Challenge in Mice“, Sci Rep, 8(1):16509
McFarlane et al. (2019) “The histone chaperone HIRA promotes the induction of host innate immune defences in response to HSV-1 infection“, PLoS Pathog, 15(3):e1007667
Mancini et al. (2019) “Wolbachia strain wAu efficiently blocks arbovirus transmission in Aedes albopictus“, bioRxiv
Creanga et al. (2020) “A comprehensive influenza reporter virus panel for high-throughput deep profiling of neutralizing antibodies“, bioRxiv


Leave A Comment