Accurate cell counts are critical in sample preparation for single cell sequencing.
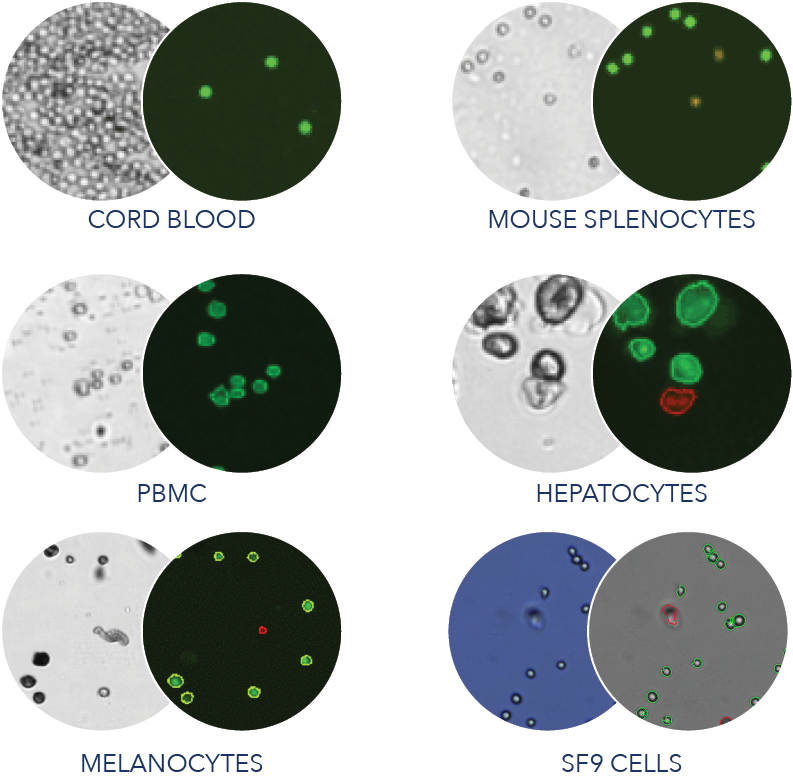
In order to establish appropriate models to study cell to cell interactions, many academic institutions have opened single cell sequencing and genomics cores. Many use transcriptome profiling together with other cutting-edge technologies to characterize how cells work within human and murine tumor microenvironments. Often core managers get a variety of cell types, at different concentrations and sizes – accuracy, consistency and being conservative with the total volume used becomes extremely important.
Cellometer guarantees accurate cell counts for your single cell sequencing.
Accurate counting and qualification of cells during the workflow of single cell sequencing is essential to determine the number of cells to load on to the microfluidic chip for analysis on the system. The typical Cellometer K2 workflow will require only 10 ul of sample. The sample is mixed 1:1 with our nuclear dyes and 20 ul of stained sample is loaded into Cellometer SD100 slides. Even then, there are times when there is simply not enough volume of cells to take 10 ul. Nexcelom has solved the problem by providing you with custom slides, SD025- which take up 4ul of stained sample. What would you do if you had a 20% margin of error for counting truly precious samples, and could complete counts in 30 seconds per sample?
Use Cellometer K2 to eliminate missteps in sample preparation.
Next-generation sequencing platforms provide a gold mine of data based on a systems approach to illuminate genotype-phenotype relationships. Eliminating missteps on sample preparation would be one simple guarantee for quality assurance and quality control. Cellometer K2 will help you achieve that balance.




Leave A Comment