Responsible for approximately one in every 18 cancer deaths in 2020, esophageal cancer (EC) is among the world’s top ten most common malignancies. Eastern Asia accounts for most of these cases, with squamous cell carcinoma comprising over 90% of all esophageal cancers in the region [1]. Exploration of candidate genes responsible for esophageal squamous cell carcinoma (ESCC) pathogenesis may provide insight into the underlying signaling pathways to uncover novel therapeutic targets.
Hou et al. previously identified thrombospondin type 1 domain-containing 7A (THDS7A) as a prominent cancer-promoting gene in ESCC, but functional downstream targets were not evaluated [2]. In this study, Jumai et al. used qRT-PCR and high-throughput screening to identify and characterize the downstream genes of THSD7A likely to promote ESCC proliferation and migration. Functional screening of genes with >40% downregulation identified 11 candidate genes expressed downstream of THSD7A.
High-throughput Screening on the Celigo Image Cytometer
Cell proliferation was measured for 11 candidate genes after shRNA transfection to identify the downstream gene(s) driving THSD7A (Figure 1). The Eca109 cells in the knockdown and control groups were transfected with lentiviruses to generate GFP-expressing cells for the proliferation measurements. Changes in the proliferation rate of the transfected cells were measured daily after initially seeding 2000 cells per well.
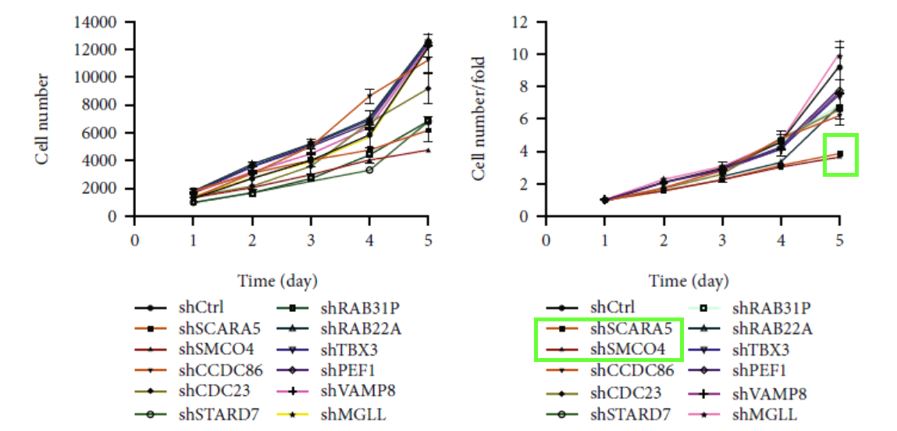
Figure 1. Cell proliferation curves for the 11 candidate genes following shRNA transfection. (Modified from Jumai et al. Journal of Immunology Research (June 2022)
The preliminary high-throughput screening identified scavenger receptor class A member 5 (SCARA5) and single-pass membrane protein with coiled-coiled domain 4 (SMCO4) as potential proto-oncogenes based on the significant reduction in Eca109 proliferation (Figure 2). The significant effects of SCARA5 and SMCO4 on Eca109 proliferation were initially determined with a mixed interference sequence. To further refine the experiment, three separate interference sequences of each gene were packaged into vectors and transfected into the Eca109 cells. The single-target experiment identified shSCARA5(PSC38272) and shSMCO4(PSC37528) as the two most significant sequences and selected these for additional in vitro functional experiments.
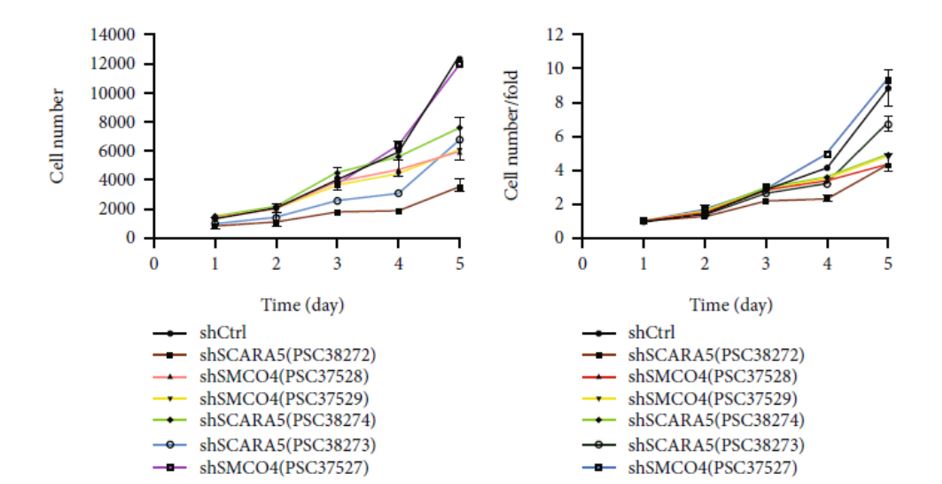
Figure 2. Cell proliferation curves for the individual interference sequences following shRNA transfection. (Modified from Jumai et al. Journal of Immunology Research (June 2022)
Additional functional verification tests confirmed that SCARA5 was the only oncogene of the 11 candidate genes originally identified. High-throughput screening on the Celigo image cytometer facilitated the investigation of a wide array of candidate genes to narrow the scope of the research quickly and efficiently to a key downstream gene involved in the pathogenesis of ESCC.
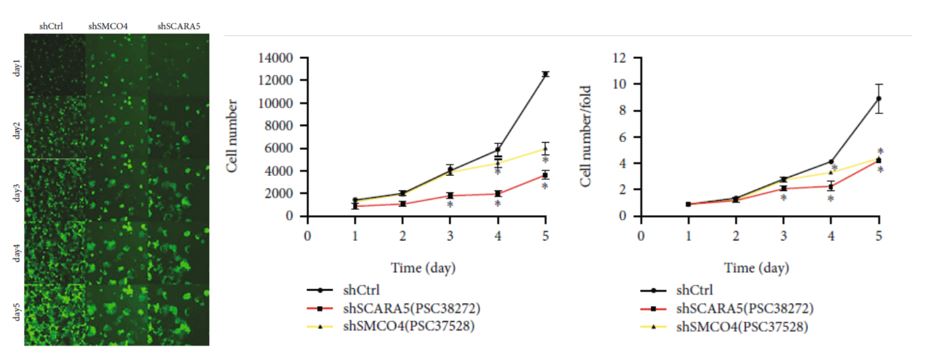
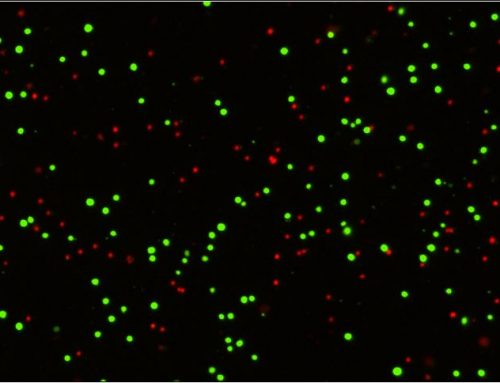
Figure 3. eGFP expression image on the Celio image cytometer used to measure cell proliferation curves for the two primary oncogene candidates. (Modified from Jumai et al. Journal of Immunology Research (June 2022)
The shRNA-SCARA5 knockdown inhibited the proliferation of the Eca109 cells. The absolute number of cells and the multiple of the change in cell number were significantly reduced compared to the control group. These results were further supported by MTT assays, clone formation experiments, scratch assays, cell cycle measurements, and apoptosis measurements that collectively implicated SCARA5 in the proliferation and migration of ESCC. It is worth noting that the Celigo can perform many of the supporting cell-based assays that were demonstrated in this study (cell cycle, apoptosis, wound healing, colony formation). Additionally, an in vivo tumorigenesis experiment demonstrated reduced tumor volume in the knockdown group supporting the cancer-promoting effects of SCARA5 observed in the in vitro experiments.
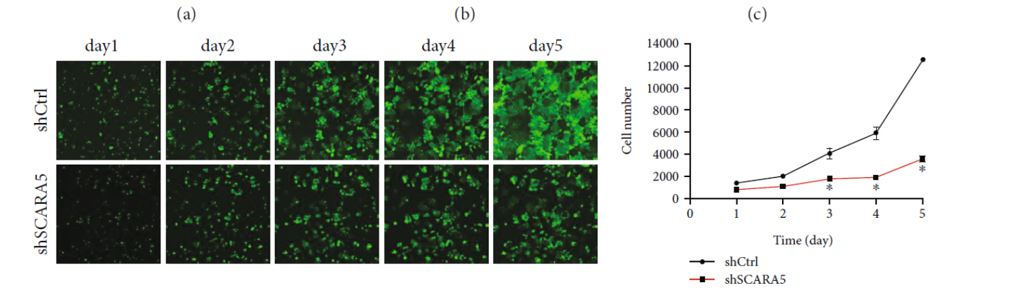
Jumai et al. then continued to probe the impact of SCARA5 in additional esophageal cancer (EC) cell lines (EC9706 and TE-1) and confirmed the cancer-promoting role of SCARA5 in EC. Finally, the group performed functional verification of SCARA5 in liver cancer (LC) cell lines to validate their methodology. Yan et al. previously reported that SCARA5 inhibits cell proliferation in LC [3] and this provides an ideal model system to validate their experimental methodology. Jumai et al. confirmed the previous findings underscoring the complexity of gene regulation across different malignancies. The application of high-throughput screening to elucidate causal relationships for specific cancers using well-established platforms, such as the Celigo, drastically reduces the time required to identify novel therapeutic targets.
The Celigo Image Cytometer offers a wide variety of applications for automated cell counting and cell-based assays with intuitive software analysis tools and automation options to accommodate high-throughput needs. Learn more about cellular models and the direct cell counting assays on our Celigo Image Cytometer Applications page or contact us directly to book a seminar or live demonstration today.
References
- Sung, H, Ferlay, J, Siegel, RL, Laversanne, M, Soerjomataram, I, Jemal, A, Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021: 71: 209- 249. https://doi.org/10.3322/caac.21660
- Hou, A. Abudureheman, L. Wang, et al., “Expression, prognosis and functional role of THSD7A in esophageal squamous cell carcinoma of Kazakh patients, Xinjiang,” Oncotarget, vol. 8, no. 36, pp. 60539–60557, 2017.
- Yan, S. Zhang, Y. Yang, et al., “Therapeutic upregulation of class A scavenger receptor member 5 inhibits tumor growth and metastasis,” Cancer Science, vol. 103, no. 9, pp. 1631– 1639, 2012.


Leave A Comment