Meeting Blood Transfusion Supply Challenges
Challenges arise with the supply and demand of RBCs for blood transfusions. The standard clinical practice of using donor-supplied RBCs is associated with significant challenges including limited donor supply, the risks of pathogen contamination, and potential alloimmunization even with standard blood compatibility testing. Mature RBCs that lack nuclei and the ability to proliferate can be generated from adult bone marrow hematopoietic stem and progenitor cells, by proliferating progenitor cells called erythroblasts. Challenges remain in the production of sufficient quantities of erythrocytes due to the limited differentiation potential of these cells
Erythrocyte Biology and Development
Erythrocytes, or red blood cells (RBCs) are formed from immature nucleated erythroblast cells and these hematopoietic stem cells form both blood and immune cells. Identifying the key growth conditions and mechanisms to achieve functional hematopoietic stem cells is a major focal point of clinical and experimental molecular therapy research.
Ectopic expression of the human BMI1 gene is often associated with the self-renewal of human hematopoietic cells. In this study1, researchers investigate the BMI1 gene’s potential to enable the extensive proliferation of functional human erythroblast cultures established from human peripheral blood mononuclear cells (PBMCs).
Finding a way to directly expand RBC precursors
In the present study (Liu et al, 2021), researchers investigated the potential of BMI1 expression to expand erythroblast progenitors in culture, ultimately for terminal differentiation and production of RBCs. The results of the study indicate that BMI1 gene expression causes expanding erythroblasts to effectively repress spontaneous differentiation. The cells are also karyotypically normal and able to terminally differentiate to RBCs when induced, providing a potential solution to blood transfusion supply challenges.
Performing high-throughput DNA synthesis-based cell proliferation assays
To aid in the accurate determination of these findings, researchers examined the proliferation of erythroblast cultures utilizing a Celigo high-throughput image cytometer. The Celigo provides rapid imaging to detect and quantify the fraction of EdU-labeled cells and DAPI-labeled cells in easily prepared microwell plates, enabling the quantification of proliferating erythroblasts.
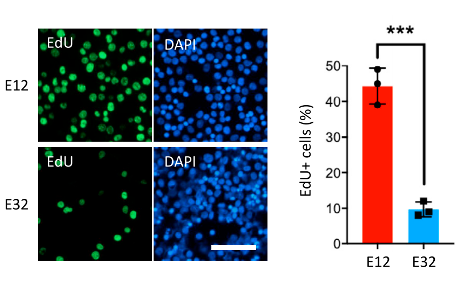
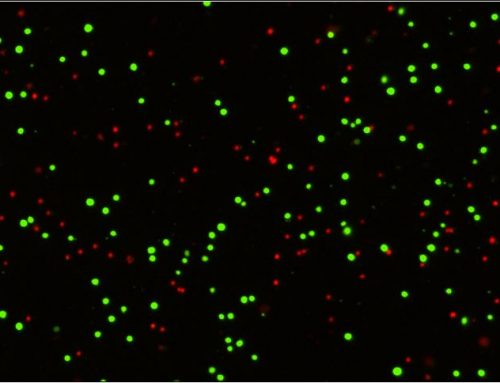
Figure 1. Cell Proliferation Assay. Shown are representative images of new DNA synthesis, assayed by the incorporation of EdU nucleotide analog, in early-stage (E12) and late-stage (E32) erythroblast cultures, during enrichment and expansion from PBMCs.
The DNA synthesis-based assay for cell proliferation was performed using EdU, a nucleotide analog marker for DNA synthesis, and 4′, 6-diamidino-2-phenylindole, dihydrochloride (DAPI). Cells were incubated in the Edu solution for four hours, fixed, and stained with anti-EdU antibody and DAPI. A proliferation index was then determined for the different cultures by quantifying the fraction of EdU-labeled cells compared to DAPI-labeled total cells using a Celigo high-throughput microwell image cytometer.
Figure 2. Example of the Celigo high-throughput image cytometer visualization tools for monitoring cell proliferation.
Reliable and accurate proliferation measurements and cell cycle studies
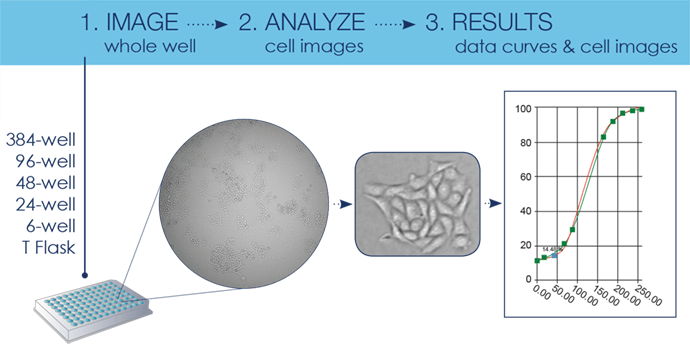
The Celigo provides an easy, reliable, and fast solution for cell proliferation and counting assays, performing direct measurements of cell numbers in each well with both adherent and suspension cell lines. The system images the entire well and counts every cell in every well, even if cells are clumped around the edges or unevenly dispersed. The Celigo simultaneously monitors the growth for every well and uses visualization tools to show quantitative growth curves for the individual wells.
Celigo proliferation assays can be an end-point or can be run in a kinetic manner, with the software conveniently grouping each timepoint under the same plate ID for easy data recall and visualization. Data can be easily exported for graphing on third-party software.

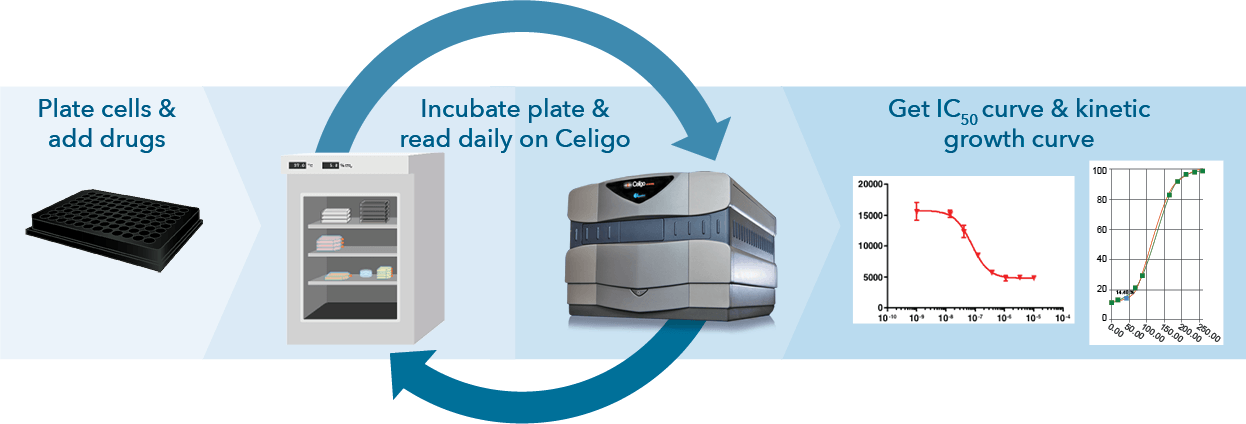
Figure 3: Celigo can be utilized for endpoint (top) or kinetic (bottom) growth studies
Direct Cell Counting and Confluence Measurements in Brightfield and Fluorescence
Cells can be directly counted in brightfield using direct cell counting or confluence applications, as well as with fluorescence, such as with Edu/DAPI. The Celigo software features a flow-cytometry-like gating interface, which can be used to segment out different cell populations based on DAPI and Edu intensity to quantify exactly how many cells are falling into the different cell cycle stages (Figure 5). Edu/DAPI for cell cycle has been performed on Celigo in several publications (1,2,3).
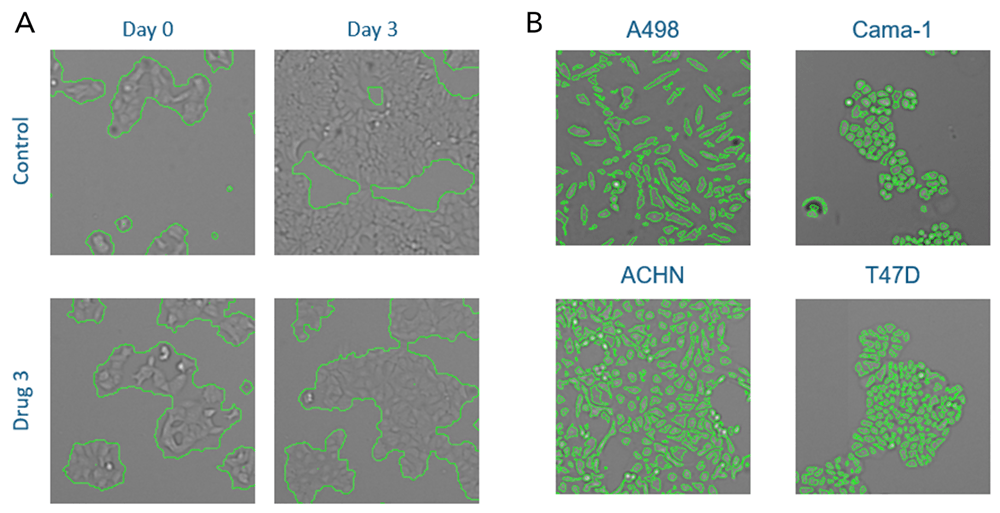
Figure 4: Cell proliferation can be directly measured in Brightfield using Confluence (A, left) or by Direct Cell Counting (B, right). Cells of different sizes and morphologies can be directly detected and quantified.
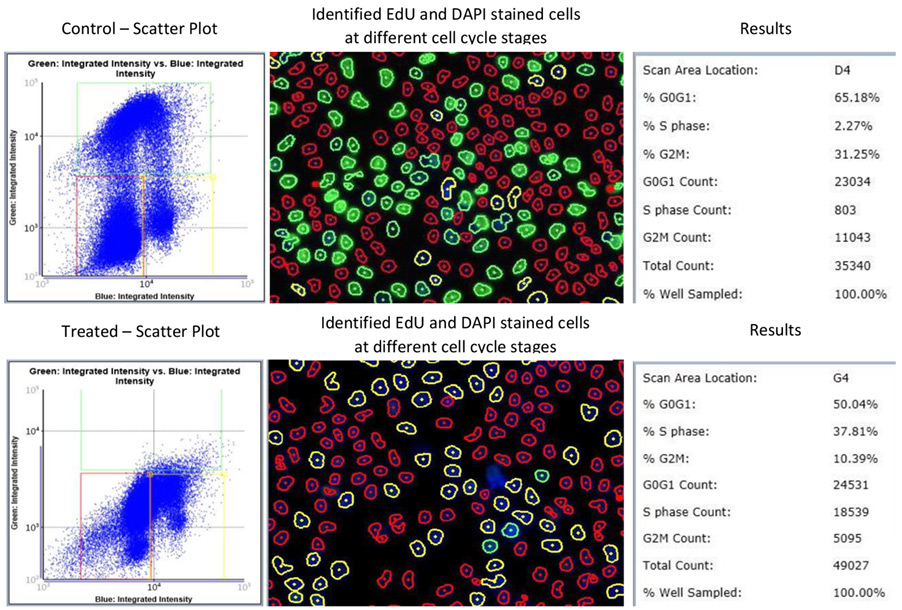
Figure 5: Celigo can conduct cell cycle and proliferation assays using Imaging Cytometry.
Versatility for fit-for-purpose research
This study demonstrates the Celigo imaging cytometer’s wide range of capabilities for cell counting, proliferation, cell cycle, and other assays for whole-well live-cell analysis and cell sample characterization in brightfield and fluorescence. Celigo increases the throughput with reliability and speed while achieving best-in-class brightfield imaging for each well to directly count cells without labeling. When applicable, fluorescence can be used to identify cell populations, such as in experiments quantifying cell viability, expression analysis, apoptosis, cell cycle, and more.
Celigo’s ability to perform simultaneous imaging and analysis allows researchers to link the instrument with automation systems, such as our stacker platform, to easily perform overnight measurements of dozens of plates for toxicity, expression, proliferation, or other kinds of studies.
References
- Liu, S., Wu, M., Lancelot, M., Deng, J., Gao, Y., Roback, J. D., Chen, T., & Cheng, L. (2021). BMI1 enables extensive expansion of functional erythroblasts from human peripheral blood mononuclear cells. Molecular therapy: the journal of the American Society of Gene Therapy, 29(5), 1918–1932. https://doi.org/10.1016/j.ymthe.2021.01.022
- Wang, YC., Morrison, G., Gillihan, R. et al. Different mechanisms for resistance to trastuzumab versus lapatinib in HER2- positive breast cancers – role of estrogen receptor and HER2 reactivation. Breast Cancer Res 13, R121 (2011). https://doi.org/10.1186/bcr3067
- Stecklein, S. R., Kumaraswamy, E., Behbod, F., Wang, W., Chaguturu, V., Harlan-Williams, L. M., & Jensen, R. A. (2012). BRCA1 and HSP90 cooperate in homologous and non-homologous DNA double-strand-break repair and G2/M checkpoint activation. Proceedings of the National Academy of Sciences of the United States of America, 109(34), 13650–13655. https://doi.org/10.1073/pnas.1203326109
- Smith KT, Sardiu ME, Martin-Brown SA, Seidel C, Mushegian A, Egidy R, Florens L, Washburn MP, Workman JL. Human family with sequence similarity 60 member A (FAM60A) protein: a new subunit of the Sin3 deacetylase complex. Mol Cell Proteomics. 2012 Dec;11(12):1815-28. DOI: 10.1074/MCP.M112.020255 Epub 2012 Sep 14. PMID: 22984288; PMCID: PMC3518139.https://www.mcponline.org/article/S1535-9476(20)33473-3/fulltext


Leave A Comment