Discovery of a Single Broad Regulator of Anti-Tumor Immunity in both T-cells and Cancer Cells
The ongoing pursuit of novel anti-cancer therapeutics must consider the potential for off-target effects on the immune system. Several anti-cancer drugs impair immune cell functionality, increasing the chance of recurrence due to the outgrowth of drug-resistant tumor cells (1). These potential off-target effects emphasize the importance of developing novel anti-cancer therapeutics that can also enhance anti-tumor immunity to create the opportunity for better patient outcomes.
CARM1 is an arginine methyltransferase responsible for the methylation of histone H3 and several other chromatin-associated proteins (2). In the present study, a team of immuno-oncologists and molecular biologists characterized the role of the CARM1 protein in both the anti-tumor functions of CD8 T-cells and the susceptibility of tumor cells to T-cell mediated killing (2). They found that inactivation of the Carm1 gene in T-cells enhanced tumor cell killing and infiltration, while inactivation of the same gene in tumor cells led to greater activation of innate immunity pathways, enhanced infiltration by CD8 T-cells and other immune cell types, as well as the overcoming of resistance to immune checkpoint inhibitors (such as CTLA-4 or PD-1) in certain cancer types.
These data point to CARM1 as a promising therapeutic target for novel therapeutics that sensitizes tumor cells to immune attack while simultaneously enhancing effector T-cell function. As described below, the Nexcelom Celigo Imaging Cytometer was used for critical in-vitro cytotoxicity assays to examine the impact of the Carm1 gene in CD8 T-cell-mediated tumor killing.
Celigo Cytotoxicity Assays
Utilizing imaging cytometry, direct cell counting of live fluorescently-labeled target cells can be performed. In comparison to other methods (such as 51Cr or LDH release) that are indirect methods of cell death, this method directly measures the killing of live target cells and can be performed in a kinetic manner.
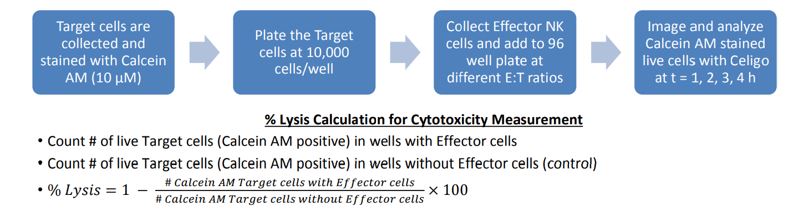
Figure 1: Standard workflow for cytotoxicity assay on Celigo. Target cells can be labeled with calcein AM or another fluorescent dye or protein, then plated on standard microwell plates and co-incubated with effector cells at different E:T (Effector: Target) ratios. Cells are imaged over time and loss of fluorescent cells is evaluated compared to control wells.
Target cancer cells are often labeled with dyes such as calcein AM or CMFDA for a simple way to fluorescently label the cells and allow the instrument to easily track their number over time. These labeled cancer cells are then co-incubated with effector cells, such as T-cells or NK cells, at different Effector: Target (E:T) ratios. As the effector cells do their work and the target cells die, they lose their fluorescent label. This loss of fluorescence is a direct marker of cell death, which can be tracked kinetically and compared to control wells with no effectors present (Figure 2). Conveniently, many experimental conditions can be tested and imaged on one standard microwell plate, allowing for ultimate flexibility in assay design.
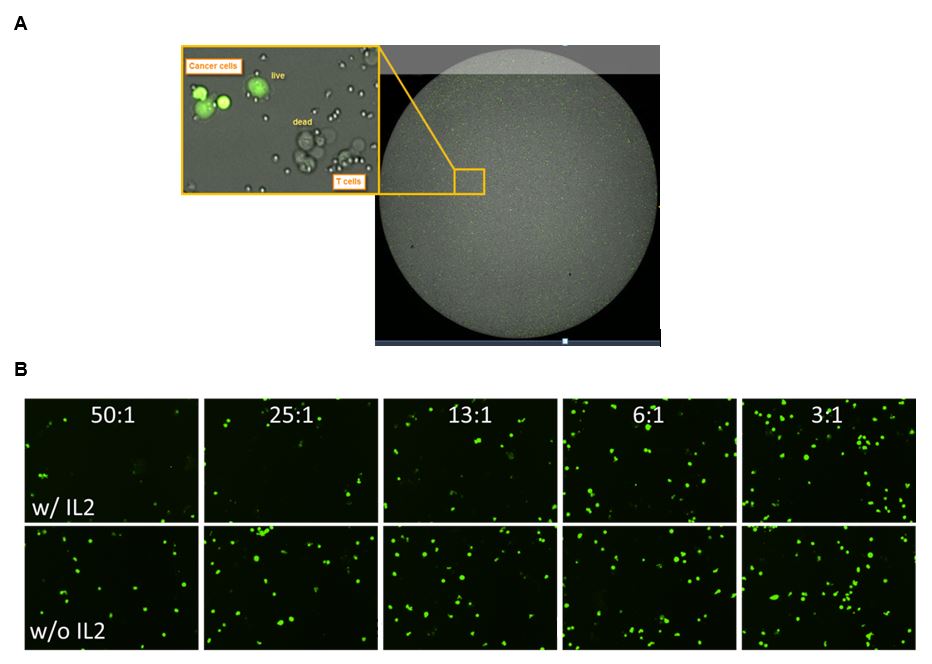
Figure 2: Standard data output from an imaging cytometry cytotoxicity assay. Panel A (top) shows an overlay of the brightfield and green channel images, with a magnified view of labeled (live) cancer cells and those that have lost their fluorescence. The smaller effector cells can also be seen in this image. Panel B (bottom) demonstrates the effect of IL2 stimulation and larger E:T ratios on the presence of labeled target cells, as there are fewer green target cells present when high numbers of IL2-stimulated target cells are present.
Data can be exported and graphed to compare different E:T ratios, kinetic timepoints, or multiple cell lines/samples, typically plotted as a percentage of the number of live cells in the control wells without effectors (Figure 3).
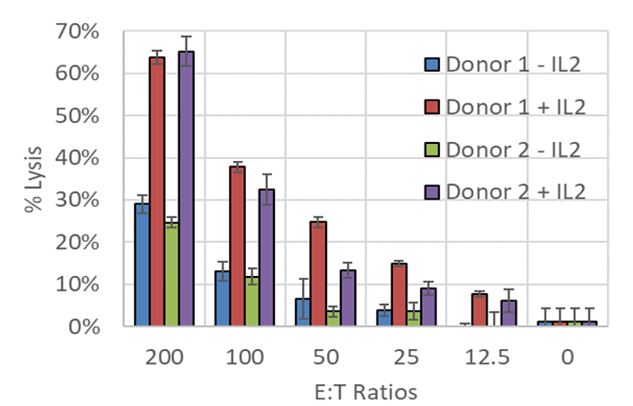
Figure 3: Celigo cytotoxicity assay data comparing effector cells from two donors with or without IL2 stimulation, and at different E:T ratios. IL2 stimulation greatly increases the killing efficiency of the effector cells from both donors.
CARM1 Inactivation in T-cells enhances anti-tumor function
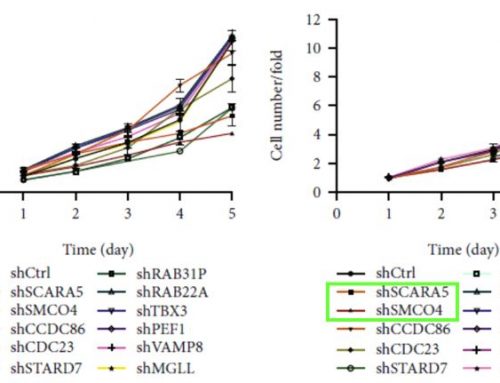
Following the identification of CARM1 as a possible negative regulator of T-cell anti-tumor function with an in-vivo CRISPR/Cas9 screen, the authors of this study proceeded to examine the effect of CARM1 by generating Cas9-edited Carm1-KO T-cells for functional assays, the first being a cytotoxicity assay performed on the Celigo instrument. Edited and control T-cells were co-incubated with ZsGreen labeled B16F10-OVA melanoma cells for 24 hours, followed by quantification of ZsGreen positive cells in each well. The results showed clear differences in cytotoxic activity, with the edited T-cells able to kill significantly more tumor cells in 24 hours (Figure 4). These data suggest that the Carm1 gene plays a role in negatively regulating T-cell immune function, leading the authors to further investigate with other experiments.
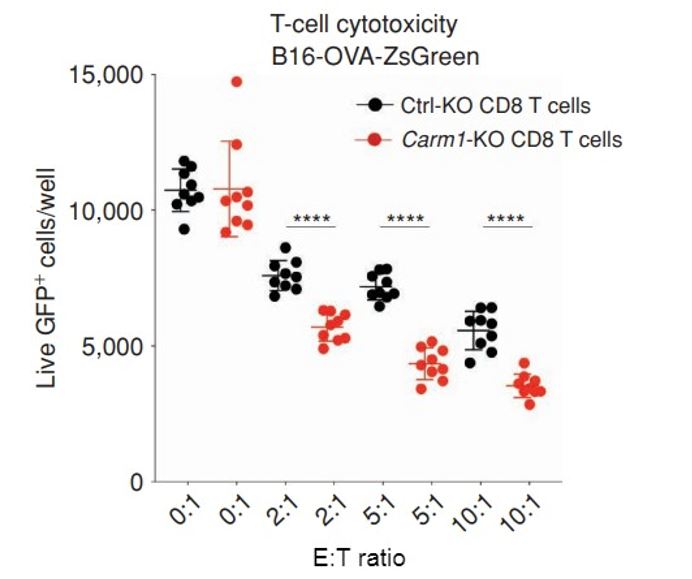
Figure 4: Inactivation of Carm1 gene in T-cells enhances tumor cell killing. B16-OVA-ZsGreen cells were seeded on 96 well plates at 5,000 tumor cells/well. Carm1-KO or control-KO CD8 T cells were then added at different effector to target (E: T) ratios. Following 24 hours of coculture, the wells were washed with PBS to remove dead tumor cells and CD8 T cells. Live, adherent tumor cells were quantified using the Celigo.
CARM1 Inactivation in Tumor Cells Enhances T-cell-mediated Anti-Tumor Immunity
After the identification of CARM1 as a potential negative regulator of T-cell antitumor function, the authors of the study examined if Carm1 inactivation in tumor cells sensitized them to T-cell attack. To address this question, they utilized several more in-vitro cytotoxicity assays using the Celigo system.
In this case, complementary labeling strategies were used; target live tumor cells were labeled with ZsGreen, or dying tumor cells were labeled with a Caspase 3/7 cell death reporter. Both approaches yielded complementary results, as Carm1 KO target cells were more susceptible to T-cell killing as measured by ZsGreen quantification of live target cells or Caspase 3/7 quantification of dying cells (Figure 5).
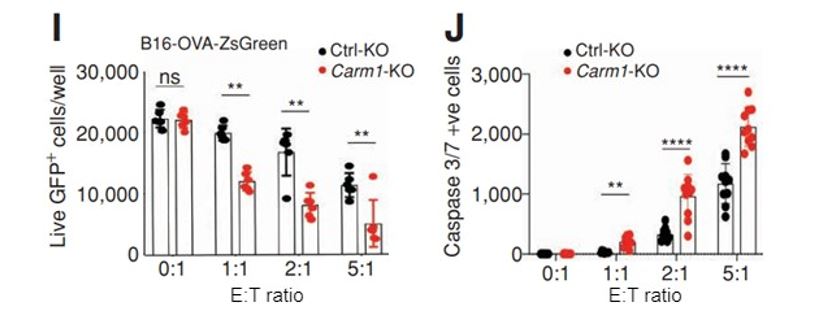
Figure 5: Inactivation of the Carm1 gene in tumor cells makes them more susceptible to T-cell attack. Carm1-KO or control-KO B16-OVA-ZsGreen cells were seeded on 96 well plates at 5,000 tumor cells/well (I). CD8 T-cells were then added at different effector to target (E:T) ratios. Following 24 hours of co-culture, the wells were washed with PBS to remove dead tumor cells and CD8 T cells. Alternatively, a caspase 3/7 reagent was added directly to the culture media (0.5 μmol/L final concentration) after coculture with ZsGreen negative cells (J). Live, adherent tumor cells or dead caspase 3/7 positive cells were quantified using the Celigo instrument.
The researchers then went further and blocked CARM1 protein function with a previously reported high affinity, small molecule inhibitor (3). Pre-treatment of two different tumor cell lines with EZM2302 sensitized them to T-cell attack, as quantified by the Celigo system (Figure 6).
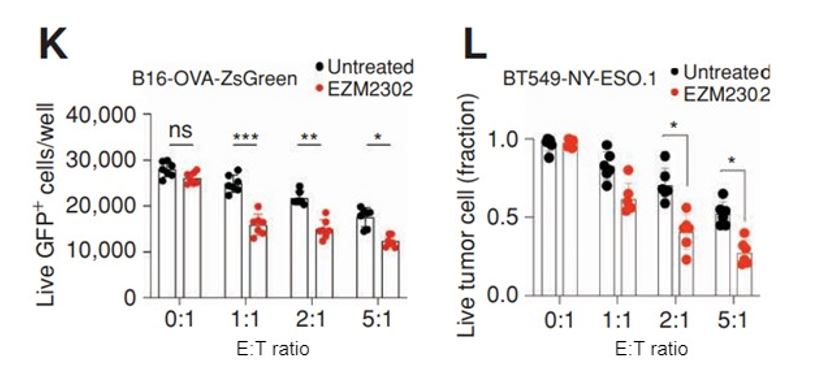
Figure 6: Blocking CARM1 protein function in tumor cells with a small molecule inhibitor makes them more susceptible to T-cell attack. B16-OVA-ZsGreen (K) or BY549-NY-ESO.1 (L) cells were seeded on 96 well plates at 5,000 tumor cells/well and some were pretreated for 24 hours with EZM2303, a Carm1 inhibitor. CD8 T cells were then added at different effector to target (E:T) ratios. Following 24 hours of co-culture, the wells were washed with PBS to remove dead tumor cells and CD8 T cells. Live, adherent tumor cells were finally quantified using the Celigo.
These data demonstrate that blocking CARM1 protein function using a small molecule inhibitor has the same effect as a Carm1 gene knockout, giving the researchers high confidence in the role of CARM1 as a negative regulator of T-cell immune function.
Using Celigo to investigate innate immune pathways downstream of CARM1
Following the discovery that CARM1 inactivation in tumor cells rendered them more sensitive to T-cell killing, the authors sought to tease out which molecular pathways were important in contributing to this phenotype. RNAseq analysis of IFNγ stimulated Carm1-KO tumor cells demonstrated Interferon-Stimulated-Genes (ISGs) were upregulated, showing that these edited tumor cells had increased responsiveness to this key cytokine secreted by activated T-cells. Further testing suggested that the cGas gene was the immune sensor downstream of CARM1 responsible for this heightened sensitivity to T-cell killing.
To test this hypothesis, the authors generated cGas-KO and Carm1-cGas-dKO cells (with both genes inactivated), which they then examined using the same in-vitro T-cell cytotoxicity assay using the Celigo system. It was found that cGas-KO cells (without Carm1 inactivated) and the double-KO were both resistant to T-cell killing (Figure 7), indicating that the cGAS enzyme is a key factor in the innate immune pathways activated in Carm1-KO cancer cells.
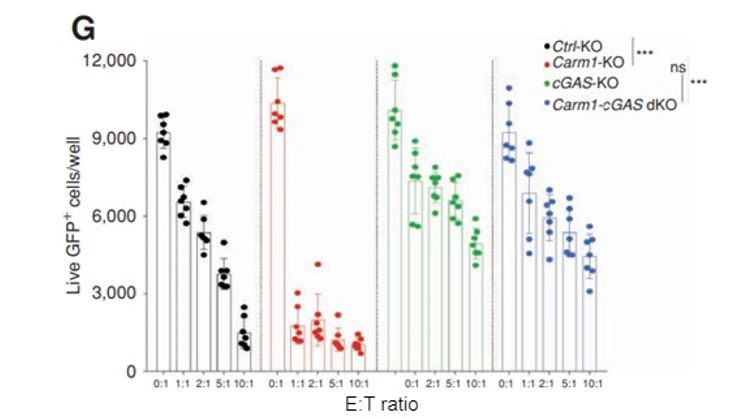
Figure 7: cGas gene inactivation in tumor cells leads to increased resistance to T-cell attack, independent of Carm1 status. Carm1-KO, cGAS-KO, Carm1-cGAS-dKO, or control-KO B16-OVA-ZsGreen cells were seeded on 96 well plates at 5,000 tumor cells/well. CD8 T cells were then added at different effector to target (E:T) ratios. Following 24 hours of co-culture, the wells were washed with PBS to remove dead tumor cells and CD8 T cells. Live, adherent tumor cells were quantified using the Celigo.
Conclusions
In this work, the authors sought to investigate the role of CARM1 in antitumor immunity utilizing a comprehensive set of experiments. In addition to the cytotoxicity assays described here, this study also utilized several traditional and cutting-edge molecular biology techniques to investigate the role of CARM1, including CRISPR/Cas9 gene editing, RNA Seq, and in-vivo screens of CD8 T-cell infiltration in tumors, among others. During the characterization of the role of CARM1 in T-cells and tumor cells, the in-vitro cytotoxicity assay from Celigo proved to be a valuable tool to demonstrate the critical importance of this protein in modulating effector T-cell function.
This is one of many examples demonstrating the utility of Celigo for in-vitro cytotoxicity assays. The ease of assay setup, coupled with the unparalleled speed of data collection and analysis, make this instrument an ideal tool to investigate T-cell cytotoxicity. If you’d like to learn more, contact us for more information or to request a live demonstration today.
References
- Bouwman, Peter, and Jos Jonkers. “The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance.” Nature reviews. Cancer vol. 12,9 (2012): 587-98. doi:10.1038/nrc3342 https://pubmed.ncbi.nlm.nih.gov/22918414/
- Wang, Lu et al. “CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis.” Cancer cell vol. 25,1 (2014): 21-36. doi:10.1016/j.ccr.2013.12.007 https://pubmed.ncbi.nlm.nih.gov/24434208/
- Drew, Allison E et al. “Identification of a CARM1 Inhibitor with Potent In Vitro and In Vivo Activity in Preclinical Models of Multiple Myeloma.” Scientific reports vol. 7,1 17993. 21 Dec. 2017, doi:10.1038/s41598-017-18446- https://pubmed.ncbi.nlm.nih.gov/29269946/


Leave A Comment