How do you accurately measure the viral neutralizing properties of antibodies?
The microneutralization assay is a powerful, high-throughput screening tool crucial to the efficient development of novel vaccines and the discovery of compounds that can prevent or treat viral disease.
A hallmark of a strong vaccine candidate is the production of high concentrations of virus-specific antibodies. While concentration, specificity, and binding characteristics are important factors, ultimately the viral neutralizing properties of the novel antibodies are critical. Here the microneutralization assay determines the relative antiviral neutralizing power (generally by inhibiting viral entry into cells) of sera from inoculated animals or patients. This is paramount in today’s context where: “Several studies have already demonstrated that individuals who recover from SARS-CoV-2 infection seroconvert and produce high levels of antibodies against the spike protein, as measured by binding assays [9,13].” However, as of the time of writing, it is unknown whether the antibodies produced by those infected by SARS-CoV-2 convey immunity and for how long.
In addition to vaccine development, the search is on for FDA approved compounds that can eliminate or reduce the duration of viral diseases. While these compounds can and will have varying mechanisms of action, the outcome sought is the prevention of viral infection, replication, or spread. With compound libraries reaching into the tens of thousands, the microneutralization assay is an efficient, biologically relevant model for antiviral activity screening.
Performing versus Optimization of Virus Neutralizing Tests
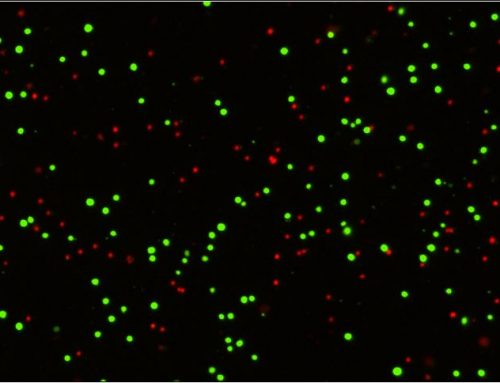
Microneutralization assays (MNA) are a derivative of the Focus Reduction Neutralization Test (FRNT) and are designed for high-throughput analysis to address the shortcomings of traditional manual readout neutralization tests. MNA are performed in 96 and 384 well formats where permissive cells are incubated with dilutions of test compounds, antibodies, or patient/animal serum, along with virus at a known infectious concentration (MOI). Following an incubation that is generally designed to allow for a single viral infection and replication cycle, the number of infected cells in each well are counted. (See figure 1 for a generalized protocol). Antibodies or compounds with antiviral neutralizing properties will result in a reduction in the number of infected cells compared to virus only conditions.

Figure 1. Generalized protocol to count the number of infected cells in each well.
Ultimately the MNA is used to determine the relative neutralizing power of a test compound expressed as IC50 and read out by enumerating the number of infected cells [1]. Determining the number of infected cells can be done by directly counting infected cells, most commonly by microscopy [1,3-7] or flow cytometry [18] following immunostaining for viral proteins such as the nucleoprotein, or by indirect methods such as bioluminescence [11,13].
Automated microscopy, such as with the Celigo Image Cytometer, can have significant advantages in optimization, speed, accuracy, and cost over other methods. With automated microscopy, it is possible to scan the entire well of each plate in under 15 minutes and directly enumerate the infected cells giving an absolute measure of the infectious viral titer (1 infected cell = 1 infectious viral particle). In comparison, flow cytometry provides a population-level measurement or percent of the population that is infected with a virus and typically requires 45 minutes for each plate. In addition to the need to detach adherent growing cells, flow instruments have difficulty providing absolute measurements of every cell in a given well. Bioluminescence based readouts can be achieved with edited viruses designed to deliver luciferase as a reporter of infection.
These constructs are most applicable to internalization inhibition assays (such a patient serum) and can produce a rapid readout. However, the luminescence readout is an indirect measure of cell number and reagent costs can be high.
Some considerations in developing and optimizing MNA include the permissive cell type being used, virus type, (wildtype, edited, or pseudotyped virus particle), and concentration (MOI)[1, 6,16].
Biological Characteristics and Available Instrumentation Impacts Type of Cell Utilized
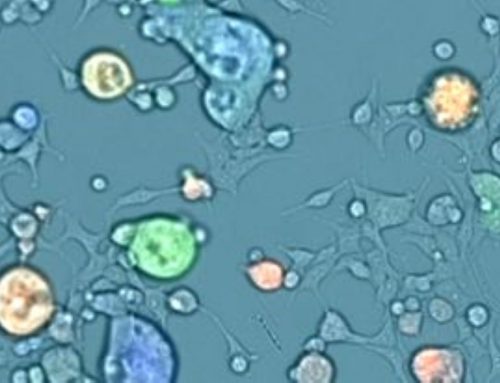
The permissive cell type used for MNA will be determined first by biology (in that the virus needs to be able to infect them) and potentially by available instrumentation. Suspension growing cells are best if the only instrumentation available for assay readout is flow cytometry. (Image cytometers such as the Celigo can enumerate both adherent and suspension growing cells as shown in this example [ href=”https://www.nexcelom.com.cn/celigo/virology-3-quantify-viral-antibody-neutralization.php” target=”_blank” countries_mode=”include” countries_input=”” countries_region=”null” cities_mode=”include” cities_input=”” cities_region=”null” states_mode=”include” states_input=”” states_region=”null” zipcodes_mode=”include” zipcodes_input=”” zipcodes_region=”null” radius_mode=”include” ]) The disadvantage of suspension growing cells is the potential loss of infected cells during wash steps.
Viral Particle Choice and Immunostaining Considerations
The choice of viral particle to be used in the assay can be determined by the process the MNA is designed to test. Live virus can be used for MNA assays that assess cell entry as well as those that test viral replication. Assays utilizing drug libraries generally necessitate the use of live virus, as compounds may target different components of the replication cycle. Here, biosafety considerations require extended fixation protocols (24 hrs. in paraformaldehyde) to render the virus inert, and detection is achieved by immunostaining.
In comparison, internalization inhibition MNA is often performed with a pseudotyped virus [3,11,16] edited to deliver a fluorescent (GFP, mKate) or bioluminescent (Luc) reporter gene. A significant advantage in the use of these pseudovirus particles is that assays can be potentially performed at lower biosafety levels, and immunostaining steps can be skipped. A potential caveat is, however, that “they do not employ authentic virus and are, therefore, always just an approximation of “real” virus neutralization. In addition, they can only be used to test entry inhibitors, but not other types of drugs. “ [9].
Determination of Optimal Multiplicity of Infection (MOI)
For an accurate neutralization assay, the amount of virus used is an important variable [9]. Here there is a balance between sampling statistics and keeping the virus titer low enough to not overwhelm the ability of the test compounds to inhibit infection [15]. In any sampling problem, the random error as a percent can be described by sqrt(n)/n *100% where n is the number of counted objects. Therefore, for a well-defined assay with less than 10% random error in the no neutralization control conditions, it is necessary to have at least a viral titer resulting in 100 infected cells. On the high end, MNA assays are generally designed for readout after a single infection cycle where each infected cell is assumed to arise from a single infectious viral particle. Therefore, viral titers should be used where the probability of two neighboring cells being infected is low (for differentiation between random infection events and re-infection events). Finally, titers should not be so high as to be insensitive to the ability of the tested article to inhibit infection or replication.
In practice, viral titers resulting in approximately 100 to 2000 infected cells/ wells are used.
References
- Shambaugh C, Azshirvani S, Yu L, et al. Development of a High-Throughput Respiratory Syncytial Virus Fluorescent Focus-Based Microneutralization Assay. Clin Vaccine Immunol. 2017 Dec 5;24(12):e00225-17. doi: 10.1128/CVI.00225-17. PMID: 29021302; PMCID: PMC5717189. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5717189/
- Swanson KA, Rainho-Tomko JN, et al. A respiratory syncytial virus (RSV) F protein nanoparticle vaccine focuses antibody responses to a conserved neutralization domain. Sci Immunol. 2020 May 1;5(47):eaba6466. doi: 10.1126/sciimmunol.aba6466. PMID: 32358170. A respiratory syncytial virus (RSV) F protein nanoparticle vaccine focuses antibody responses to a conserved neutralization domain
- Daniloski Z, Guo X, Sanjana NE. The D614G mutation in SARS-CoV-2 Spike increases transduction of multiple human cell types. bioRxiv [Preprint]. 2020 Jun 15:2020.06.14.151357. doi: 10.1101/2020.06.14.151357. PMID: 32587969; PMCID: PMC7310625. The D614G mutation in SARS-CoV-2 Spike increases transduction of multiple human cell types
- Elicitation of broadly protective immunity to influenza by multivalent hemagglutinin nanoparticle vaccines S Boyoglu-Barnum, D Ellis, RA Gillespie… – BioRxiv, 2020 – biorxiv.org (Celigo)
- White KM, Abreu P Jr, et al. Broad Spectrum Inhibitor of Influenza A and B Viruses Targeting the Viral Nucleoprotein. ACS Infect Dis. 2018 Feb 9;4(2):146-157. doi: 10.1021/acsinfecdis.7b00120. Epub 2018 Jan 4. PMID: 29268608; PMCID: PMC6145453. (Celigo)
- Rosen O, Chan LL, et al. A high-throughput inhibition assay to study MERS-CoV antibody interactions using image cytometry. J Virol Methods. 2019 Mar;265:77-83. doi: 10.1016/j.jviromet.2018.11.009. Epub 2018 Nov 20. PMID: 30468747; PMCID: PMC6357230. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6357230/ (Celigo)
- Gordon DE, Jang GM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 Jul;583(7816):459-468. doi: 10.1038/s41586-020-2286-9. Epub 2020 Apr 30. PMID: 32353859; PMCID: PMC7431030. https://www.nature.com/articles/s41586-020-2286-9 (Celigo)
- Sirivichayakul C, Sabchareon A, et al. Plaque reduction neutralization antibody test does not accurately predict protection against dengue infection in Ratchaburi cohort, Thailand. Virol J. 2014 Mar 12;11:48. doi: 10.1186/1743-422X-11-48. PMID: 24620925; PMCID: PMC3975240. (PRNT does not predict protection against dengue) (manual readout)
- Amanat F, White KM, et al. An In Vitro Microneutralization Assay for SARS-CoV-2 Serology and Drug Screening. Curr Protoc Microbiol. 2020 Sep;58(1):e108. doi: 10.1002/cpmc.108. PMID: 32585083; PMCID: PMC7361222. (Celigo)https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7361222/
- Riva L, Yuan S, et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020 Jul 24. doi: 10.1038/s41586-020-2577-1. Epub ahead of print. PMID: 32707573. (Celigo) https://www.nature.com/articles/s41586-020-2577-1_reference.pdf
- Carnell GW, Ferrara F, et al. Pseudotype-based neutralization assays for influenza: a systematic analysis. Front Immunol. 2015 Apr 29;6:161. doi: 10.3389/fimmu.2015.00161. PMID: 25972865; PMCID: PMC4413832. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4413832/ info about pseudotype virus particles for influenza serum screens virus can deliver luminescence.
- https://www.biorxiv.org/content/10.1101/2020.07.23.212357v1
- Mercado NB, Zahn R, et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020 Jul 30. doi: 10.1038/s41586-020-2607-z. Epub ahead of print. PMID: 32731257. https://www.nature.com/articles/s41586-020-2607-z_reference.pdf MNA by luciferase.
- https://www.nexcelom.com/applications/virology/antibody-neutralization/cytomegalovirus-neutralization-assay-using-fluorescence/
- https://www.virology.ws/2009/05/28/influenza-microneutralization-assay/
- Almasaud A., Alharbi N.K., Hashem A.M. (2020) Generation of MERS-CoV Pseudotyped Viral Particles for the Evaluation of Neutralizing Antibodies in Mammalian Sera. In: Vijay R. (eds) MERS Coronavirus. Methods in Molecular Biology, vol 2099. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0211-9_10 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7123069/
- Okba NMA, Müller MA, et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerg Infect Dis. 2020 Jul;26(7):1478-1488. doi: 10.3201/eid2607.200841. Epub 2020 Jun 21. PMID: 32267220; PMCID: PMC7323511. (immunospot reader)
- Baldwin WR, Livengood JA, et al. Purified Inactivated Zika Vaccine Candidates Afford Protection against Lethal Challenge in Mice. Sci Rep. 2018 Nov 7;8(1):16509. doi: 10.1038/s41598-018-34735-7. PMID: 30405178; PMCID: PMC6220238.
- https://www.nexcelom.com.cn/celigo/virology-3-quantify-viral-antibody-neutralization.php


Leave A Comment