Note: Figures referenced in this review are available to view in the original manuscript “A SARS-CoV-2 protein interaction map reveals targets for drug repurposing”
A Novel Virus Emerges
In December 2019, the Wuhan Municipal Health Commission in China reported an unusual cluster of pneumonia cases. This outbreak was ultimately linked to a novel coronavirus termed SARS-CoV-2 based on its similarity with the coronavirus responsible for 8000 cases of severe acute respiratory syndrome (SARS) in 2003. There is a close phylogenetic relationship between the infectious agents responsible for SARS and what would eventually be named COVID-19; but they are very different diseases. The SARS epidemic was tamped out relatively quickly, while COVID-19 now rages on as a pandemic.
SARS-CoV-2 virus with the spike proteins expressed on the surface, which can infect humans causing Covid-19.
Despite a draconian months-long lockdown of Wuhan that began January 23, the highly infectious genie had already escaped the lamp. At the writing of this post 6 months after the initial report, the number of novel coronavirus cases has surpassed 9 million, with more than 470,000 deaths worldwide. Unlike influenza that wanes in the warm months, COVID-19 infections show no signs of abating. As we look toward a long summer of steady new cases (plateaued at 20,000/day in the U.S. for 30 days), researchers are urgently working to identify and test prophylactics and treatments that can prevent more deaths until an effective vaccine is available, hopefully in early 2021.
Until the Vaccine
Between preventative vaccines and palliative treatment for the sickest COVID-19 patients lies the realm of existing drugs that could be repurposed for antiviral prophylaxis. The Nature article “A SARS-CoV-2 protein interaction map reveals targets for drug repurposing” describes how Gordon et al. identified and tested these compounds. Working from the early publication of the viral genome, more than 100 researchers around the world collaborated to clone, tag, and express proteins encoded in the virus’ RNA genome in human cells. Next, binding between viral and human proteins was assessed to build an interactome of protein-protein interactions (PPIs). Gene Ontology (GO) term enrichment indicated which major cell processes were likely affected and revealed 66 druggable targets for 69 existing compounds. A subset of these were assessed for antiviral activity with two assays in laboratories in Paris and New York. The aggregated results indicate that two sets of agents have potential: mRNA translation inhibitors and Sigma receptor regulators.
Understanding Host-Virus PPIs
The first step was to develop a systematic map of the interactions between host and viral proteins. The researchers cloned, tagged, and expressed 26/29 (89.6%) viral proteins in the human kidney cell line HEK293T/17. Affinity-purification mass spectrometry (Fig. 1) identified 332 PPIs (Figs. 2-4). Interestingly, among 10 other viruses, the host-protein interaction pattern was most similar to those for West Nile Virus and Mycobacterium tuberculosis, which also preferentially infect neurons and lung cells, respectively.
Cellular Pathways Affected by COVID-19
Detailed GO analyses of the interactome shed new light on relationships between viral and host proteins, with effects on DNA replication, epigenetic and gene expression regulation, lipid modification, nuclear transport, mitochondria, and the extracellular matrix (Fig. 3). Viral proteins also interact with several innate immune pathways (e.g., interferon, nuclear factor-kB, and ubiquitin ligases), host translational machinery, and bromodomain proteins involved in epigenetic regulation (Fig. 4). All of these were potential targets for existing drugs that could be co-opted to tamp down the infection, but extensive in vitro testing was needed to assess their efficacy.
Drug Repurposing
The authors used two approaches to find drugs that could modulate the 332 PPIs: 1) chemoinformatic analysis of open-source chemical databases and 2) a target- and pathway-specific literature search. From more than 10,000 possibilities, the list was winnowed down to 69 based on FDA approval status, activity better than 1 mM, and commercial availability. The final list included 29 approved drugs, 12 investigational new drugs, and 28 preclinical candidates.

Currently FDA approved drugs can be repurposed to test efficacy against Covid-19.
Do Any Inhibit Viral Activity?
Next, 48/69 (69.5%) of these drugs and compounds and an additional 18 were subjected to antiviral activity assays at two institutions (Fig. 6 and Extended Data Fig. 8-9). Mt Sinai Hospital in New York employed a medium-throughput immunofluorescence-based assay to detect the viral NP protein using the Celigo Image Cytometer from Nexcelom Bioscience. Human kidney cells were treated with compounds for 2 h before the virus was added, and the cells were incubated for 48 h before they were fixed and fluorescently labeled for the viral NP protein and counterstain with DAPI for total cells. Infectivity was measured by viral NP protein accumulation and expressed as the population of infected cells. At the Institut Pasteur in Paris, researchers monitored viral RNA levels in cell supernatant with quantitative reverse-transcription polymerase chain reaction; plaque assays were performed in parallel. Viral growth and compound cytotoxicity were measured at both locations. For a subset of compounds, infectious viral titers in cell supernatants were measured using the Median Tissue Culture Infectious Dose (TCID)50 method, and time course assays with drug addition at different times relative to viral infection (-2, 0, +2, and +4 hours from infection).
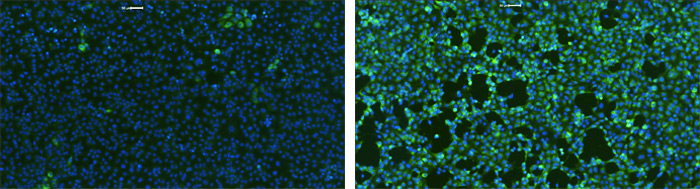
Example SARS-CoV-2 infected host cells labeled with DAPI and AF488 fluorescence to analyze the infection rate after treatments.
The results revealed that two molecule classes had the strongest effect: protein biogenesis inhibitors (zotatifin, ternatin-4, and PS3061) and Sigma1 and -2 receptor ligands (haloperidol, PB28, PD-144418, and hydroxychloroquine [HCQ]). PB28, zotatifin, and HCQ decreased viral NP protein levels, suggesting that they exert antiviral effects before the virus exits the cell. They also inhibited NP expression up to 4 h after infection. The authors concluded that these compounds act on viral replication.
- PB28 is an experimental anticancer compound that was found to be 20 times more potent against SARS-CoV-2 than HCQ.
- Zotatifin is currently being evaluated in a Phase I/II clinical trial of patients with solid tumors. It inhibits elongation factor-1A (eEF1A) that unwinds complex RNA structures, hampering translation elongation needed to produce adequate levels of viral proteins.
- HCQ is an immunomodulatory drug used for 60 years to treat malaria and autoimmune conditions. Although it showed antiviral activity, its safe use is limited by off-target effects that may induce cardiac adverse drug reactions.
Discussion
A host-directed intervention can be a useful stop gap before vaccine distribution. The Nature article by Gordon et al. is impressive for its scale and velocity. It represents an early attempt to identify possible COVID-19 treatments in the existing pharmacologic armamentarium and underscores that medium- and high-throughput assays are necessary to quickly test compounds that may be useful in the battle against this raging pandemic.
A total of 69 compounds were initially identified. Of these, 48 and an additional 18 (n=66) were tested for antiviral activity. It is not surprising that protein biogenesis inhibitors (zotatifin, ternatin-4, and PS3061) exerted antiviral effects, given that viruses are dependent on host machinery for their replication and propagation. They are also effective at concentrations of 10-100 nM, making them attractive candidates for human use.
The role of Sigma receptors is somewhat muddier. These non-opioid receptors bind several classes of psychotropic drugs, but their molecular mechanisms are unclear. It has been argued that they are not traditional receptors; rather, they are unique integral membrane chaperones or scaffolding protein that can be pharmacologically regulated. Gordon and colleagues hypothesized that they may decrease infectivity by perturbing the cell stress response. A review by Su et al. stated that stimulation of Sigma 1 by agonists or stressors induces it to translocate to the cell membrane where it interacts with ion channels, receptors, and kinases. Two of the tested drugs (clemastine and cloperastine) are approved antitussives and antihistamines and have been used for decades. Still, the study authors note that it is unclear if their pharmacokinetics are suitable for antiviral use as they also bind to side-effect targets. This is also the issue with HCQ, which increases the risk of cardiac arrhythmia. Indeed, on June 15, 2020, the U.S. FDA revoked its emergency use authorization for HCQ, stating that the “known and potential benefits…no longer outweigh the known and potential risks for authorized use.” Clearly, much more work is needed to untangle the role of Sigma receptors in SARS-CoV-2 infection.
There is one major limitation of this study that should be addressed in future work. Although HEK293T/17 cells are infected by SARS-CoV-2, the primary site of infection is the lung. Establishing assays in a more relevant cell line are needed before moving promising compounds to animal trials.
Still, the rapidity of this work (prepublished just 4 months after the first outbreak) demonstrates the utility of this experimental pipeline for drug discovery to test pan-viral strategies. It is possible that host factor-targeting agents could be used in combination with other drugs that inhibit viral enzymes. Medium- to high-throughput screening will hopefully hasten the development of effective antivirals and combination therapies for SARS-CoV-2/COVID-19.
Reviewed article:
Gordon DE, Jang GM, Bouhaddou M, et al. A SARS CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020 [prepublication]. doi: 10.1038/s41586-020-2286-9
Further reading:
Best R, Boice J. Where the latest COVID-19 models think we’re headed – and why they disagree. Available at: https://projects.fivethirtyeight.com/covid-forecasts/
Cyranoski D. Profile of a killer: the complex biology powering the coronavirus pandemic. Available at: https://www.nature.com/articles/d41586-020-01315-7
Kim FJ. Introduction to sigma proteins: evolution of the concept of sigma receptors. Handb Exp Pharmacol 2017;244:1-11.
Shippey EA, Wagler VD, Collamer AN. Hydroxychloroquine: an old drug with new relevance. Cleve Clin J Med 2018;85(6):459-467.
Su TP, Su TC, Nakamura Y, Tsai SY. The sigma-1 receptor as a pluripotent modulator in living systems. Trends Pharmacol Sci 2016;37(4):262-278.
U.S. FDA. COVID-10 Update: FDA revokes emergency use authorization for chloroquine and hydroxychloroquine. Available at: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-chloroquine-and
Zimmer C. Old drugs may find a new purpose: fighting the coronavirus. Available at: https://www.nytimes.com/2020/04/30/health/coronavirus-antiviral-drugs.html
Coronavirus tracking:
https://www.worldometers.info/coronavirus/
http://91-divoc.com/pages/covid-visualization/

Author Lindsay Reese, PhD, ELS
Dr. Reese is a bench-trained neuroscientist with 10 years of scientific communications experience. She obtained a B.S. in Biopsychology & Cognitive Science from the University of Michigan, completed her doctoral dissertation in Neuroscience & Cell Biology at the University of Texas Medical Branch, and studied neurodegeneration as a postdoctoral fellow at Oregon Health & Science University.

Co-Author Dr Leo Chan, PhD
Dr Leo Chan currently serves as the Technology R&D Manager at Nexcelom Bioscience LLC, Lawrence, MA. His research involves in the development of instrument and applications for the Celigo and Cellometer image cytometry system for detection and analysis of cells for oncology and immunology research. He is a member of the American Association of Cancer Research and the American Association of Immunologists. He received his B.S., M.S., and Ph.D. in Electrical and Computer Engineering from the University of Illinois at Urbana-Champaign (2000-2008).



Leave A Comment