The current state of SARS-CoV-2 research
The COVID-19 pandemic has increased SARS-CoV-2 research globally. The full-throttle race to develop novel therapeutics and effective antivirals against the virus resulted in three publicly available vaccines from Moderna, Pfizer (mRNA-based), and Johnson & Johnson (viral vector-based). SARS-CoV-2 RNA genome is approximately 30 kilobases long, making it one of the largest viral genomes, creating challenges in studying the virus using reverse genetics. Generating infectious viruses currently requires researchers to employ unconventional methods atypical for traditional RNA viruses as two components are needed to be present in the host eukaryotic cell to generate or rescue the infectious virus.
- The genetic material of the virus must be encoded in a yeast artificial chromosome (YAC) or bacterial artificial chromosome (BAC) and transfected into the cell.
- The nucleocapsid of the virus needs to be expressed in trans (1, 2). These systems have been successfully developed to study the pathogenicity of the virus in vitro and in vivo using reporters.
What is reverse genetics?
There are two types of genetic research employed for the molecular study of an organism. Forward genetics looks to identify the gene(s) responsible for a specific phenotype or physical trait. The opposite is true for the second type, reverse genetics (RG), where the function of a gene is studied by making changes in its sequence and observing how these changes impact the organism physically or at the molecular level. This genetic analysis is possible thanks to advances in recombinant DNA technology. RG involves the manipulation of the genetic material usually through site-directed mutagenesis, gene deletion, and transposon mutagenesis to elicit a phenotype that would inform the researcher on the gene function (3).
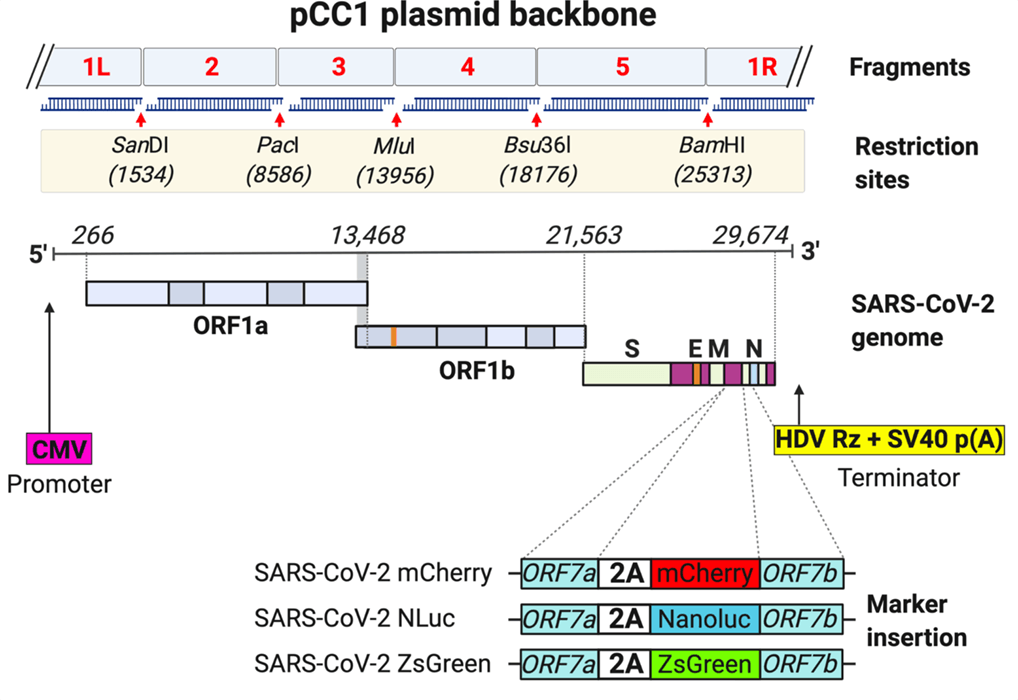
Figure 1. Schematic representation of plasmid with icDNA of SARS-CoV-2
Simplifying the study of SARS-CoV-2 and variants
With an increased focus on the pandemic, whole genomes of SARS-CoV-2 and several of its clinical variants are becoming widely available to the public every day (4, 5). This vital information can assist researchers in developing a simplified RG approach for constructing a synthetic plasmid resembling the genome of the virus. As described in Rhin.S. et.al in their most recent article, one method uses a plasmid system to rescue the infectious virus in the host cell by simply transfecting the plasmid with the infectious cDNA (icDNA) (6). This simple method grants different laboratories that previously had not worked with coronavirus the capacity to produce infectious clones of SARS-CoV-2 from a single transfection. Additionally, this genetic system allows the introduction of fluorescent reporters like mCherry, ZsGreen, or Nanoluciferase (NLuc) for rapid detection of the virus.
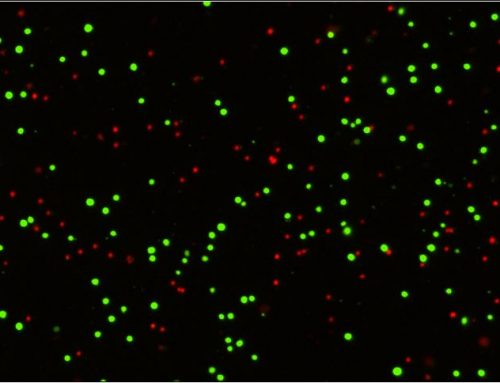
Figure 2: Plaque assays of SARS-CoV-2 expressing mCherry and ZsGreen. After 5 passages mutations resulting in a non-fluorescent virus has taken over the culture in 1 out of 3 replicates.
Combining reverse genetics with image cytometry to identify mutations and viral plaques
This RG system for COVID-19 research development employed the use of the Celigo high-throughput image cytometer to detect and quantify the percent of fluorescent plaques. This assisted in the determination that in the majority of the cases, the fluorescent virus appears to remain stable without introducing mutations. Once a mutation occurs, in this case, loss of fluorescence, this strain will take over the culture. Another advantage this RG tool brings to the field is the ability to easily generate and validate antibodies against the structural and nonstructural proteins of the virus. Using traditional biochemical techniques including western blot, immunoprecipitation, and immunofluorescence, the antibodies against these viral proteins showed to be effective at staining the infected cells and identifying protein-protein interactions (6). This approach helped identify the expression of the ORF10 protein previously thought to be a misannotated gene that does not produce a protein (7).
Adaptation for high-throughput development
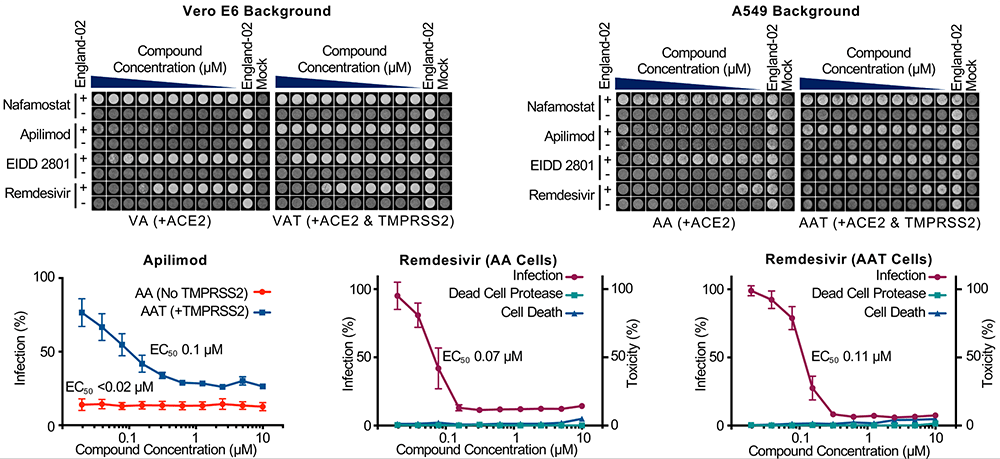
Research efforts in the last year have shown that SARS-CoV-2 can infect several cell types (8) and requires two factors to effectively infect human cells, angiotensin-converting enzyme 2 (ACE2) and TMPRSS serine protease. ACE2 is the main receptor and TMPRSS will cleave the viral S protein, priming the virus for infection. Using traditional lentiviral transduction techniques, a cell line can express each factor independently or in tandem so both are readily available to the scientific community. These cell lines can easily be adapted to medium and high-throughput formats for the screening of antiviral compounds, development of vaccine candidates, and determining the mechanism of infection.
The Celigo image cytometer was used to score cytopathic effects in response to the drug Apilimod compared to Remdesivir. This system has multiplex capabilities allowing for the determination of the effective dose of the drug (EC50) as well as the viability of the cells to assess the toxicity of the compound. The results show Apilimod was successful at reducing the infectivity of the virus in cells expressing only ACE2 but not TPRMSS2 (6). This result highlights effective tools able to effectively determine the mechanism of infection as well as the role many host factors play during infection.
Demonstrating the effectiveness of this toolkit, the permissive cell lines were used to isolate three clinical isolates (CVR-GLA-1, CVR-GLA-2. CVR-GLA3). The results show that the growth of these clinical isolates was enhanced in the permissive cell lines expressing ACE2 and TMPRSS. Additionally, antibodies generated with this toolkit were able to specifically bind to the target protein in the clinical isolate. This further validates the reagents created to study clinical isolates and further important research(6).
Figure 3: Well clearance assay. The Celigo image cytometer was used to score the cytopathic effect in response to Apilimod. Cells expressing only ACE2 +Apilimod were resistant to the infection. However, upon expression of ACE2 and TMPRSS2 the cells became permissive. EC50 was calculated from data captured on the Celigo image cytometer.
Overall, this RG tool kit consists of three parts: 1) plasmid with viral icDNA, 2) antibodies against structural and non-structural proteins, and 3) permissive cell lines to study the mechanism of infection that are available to the community. Thus, providing the virology field an additional tool to quickly characterize new viral strains and assess the effectiveness of small compounds on viral infectivity.
References
- T. Thi Nhu Thao et al., Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature. 582, 561–565 (2020).
- C. Ye et al., Rescue of SARS-CoV-2 from a Single Bacterial Artificial Chromosome. MBio. 11 (2020), doi:10.1128/mBio.02168-20.
- A. J. Griffiths, J. H. Miller, D. T. Suzuki, R. C. Lewontin, W. M. Gelbart, Reverse genetics (2000).
- D. Licastro et al., Isolation and Full-Length Genome Characterization of SARS-CoV-2 from COVID-19 Cases in Northern Italy. J. Virol. 94 (2020), doi:10.1128/JVI.00543-20.
- H. Wang et al., The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1629–1635 (2020).
- S. J. Rihn et al., A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research. PLoS Biol. 19, e3001091 (2021).
- D. Kim et al., The Architecture of SARS-CoV-2 Transcriptome. Cell. 181, 914-921.e10 (2020).
- Y. J. Hou et al., SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 182, 429-446.e14 (2020).


Leave A Comment