SARS-CoV-2 Variants necessitate constant monitoring of neutralization activity
The severe acute respiratory coronavirus 2 (SARS-CoV-2) has caused the ongoing COVID-19 pandemic, infecting more than 206 million people and causing upwards of 4.34 million deaths at the time of writing. Vaccine development in 2020 was swift and led to the availability of multiple vaccines currently in use around the world. Most of these vaccines target the Spike protein of the virus due to its critical role in recognizing the ACE2 receptor and entering host cells (1).
These vaccines are highly effective at preventing severe illness and death from SARS-CoV-2. Unfortunately, the emergence of SARS-CoV-2 ‘Variants of Concern’ (VOC) has necessitated monitoring the efficacy of current vaccines administered around the world, as many of these VOCs carry Spike protein polymorphisms that may enable escape from neutralizing antibodies produced by our current vaccines (2).
The Sputnik V vaccine, an adenoviral vector vaccine developed at the Gamaleya National Research Centre for Epidemiology and Microbiology in Moscow, Russia, is currently approved for use in Russia, Argentina, Belarus, Hungary, and over 45 other countries. Based on an interim analysis of phase 3 trials held in Russia from September – November 2020, this vaccine is 91.6% effective against infection (3). However, many VOCs with Spike protein polymorphisms were either not present or substantially transmitting in Russia during this period.
In the present study, Ikegame et al at the Icahn School of Medicine at Mt. Sinai sought to evaluate the neutralization activity of sera from Sputnik V recipients in Argentina, against different Spike protein polymorphisms. Our Celigo Imaging Cytometer was used extensively in these experiments, as seen below.
Development of a VSV-Spike surrogate assay
To assess the neutralization activity of sera from Sputnik V recipients without the use of intact SARS-CoV-2 virus, which would necessitate BSL3 facilities, Ikegame et al instead utilized an assay involving replication-competent vesicular stomatitis virus (rcVSV) as a surrogate virus expressing the SARS-CoV-2 Spike instead of VSV-G. Syncytia formed by transfected rcVSV-CoV2-S were imaged in brightfield, while rcVSV-CoV2-S also carried an EGFP reporter to allow for easy visualization of virally infected cells using the Green channel on the Celigo (Figure 1).
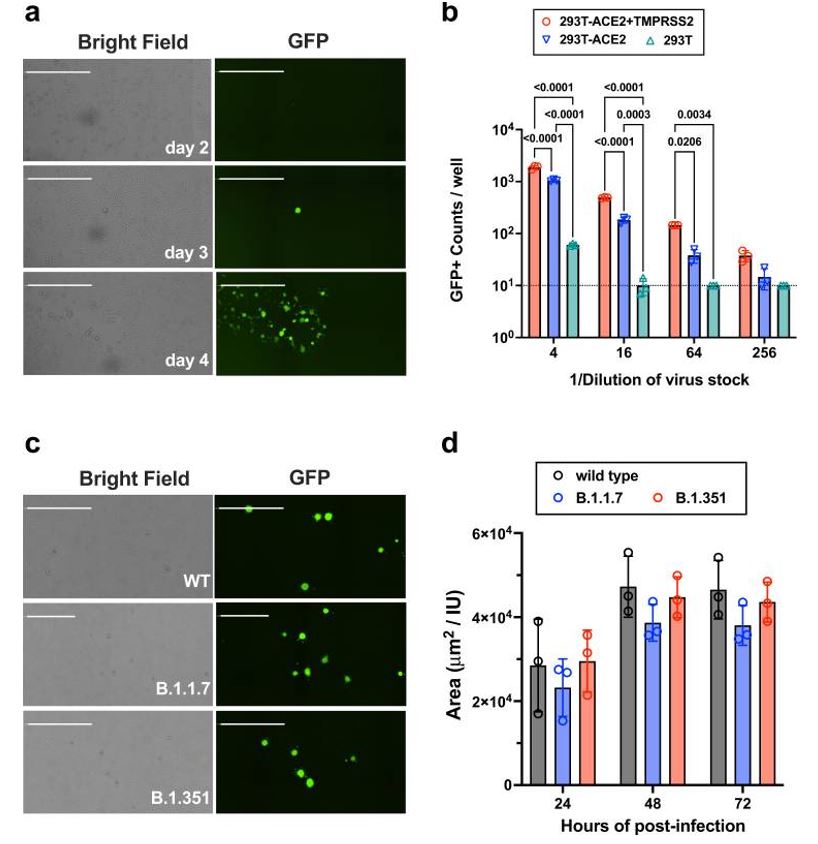
Figure 1: Titration and infection kinetics of rcVSV-CoV2-S. Panels a.) and c.) show representative images of infection, either kinetically in a.) or by different rcVSV-CoV2-S constructs carrying different spike protein polymorphisms in c.). Panel b.) compares entry efficiency of different virus stock dilutions onto the parental 293T cell line or more permissive derivations, while panel d.) shows similar infection kinetics between the different rcVSV-CoV2-S constructs. Figure and text adapted from Ikegame et al, 2021.
This initial quantification of rcVSV-CoV2-S infection, utilizing constructs carrying the different Spike protein polymorphisms, was necessary to ensure that infection kinetics were not significantly different between the rcVSV-CoV2-S constructs.
Celigo Imaging Cytometer provides rapid imaging and analysis for microneutralization assays
The Celigo Imaging Cytometer has been established as an invaluable tool for virology researchers, due to the rapid whole well imaging of microwell plates and advanced quantification of total and cell subpopulations across the entire well surface.
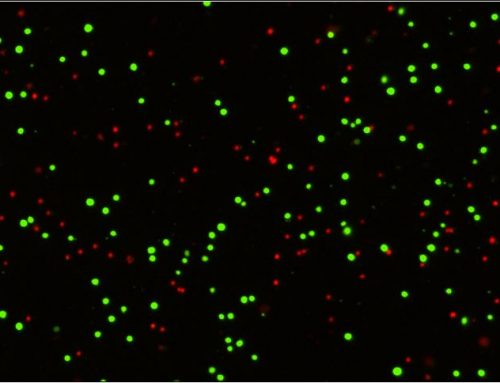
Microneutralization assays to evaluate the neutralization activity of sera against a virus can be achieved by having a serial dilution of serum across many wells on a plate containing cells infected with the virus. Total cells can be labeled with DAPI while virus-infected cells can be detected with antibody staining or a fluorescent reporter, such as GFP. The software can then determine how many total cells are infected by counting the number of GFP-positive cells against total cells, and the researcher can evaluate if more concentrated serum dilutions are reducing the number of GFP-positive cells (Figure 2).
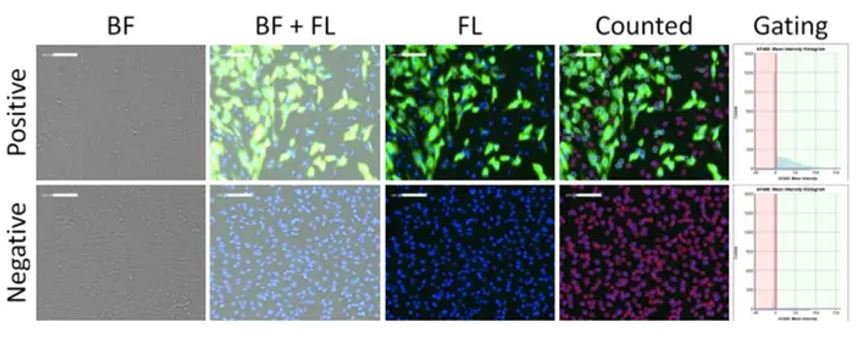
Figure 2: Brightfield and fluorescent images of viral infection, and fluorescence intensity gating of positive and negative infection of CRFK cells using optimized scanning and analysis settings from image cytometry. In this analysis, DAPI nuclei are used as a mask to count the total objects (shown in “counted”), and GFP +/- populations can be gated accordingly in the Celigo software. Figure adapted from Pearson et al (4).
To characterize the neutralization activity of sera from Sputnik V vaccine recipients, Ikegame et al utilized the Celigo to quantify GFP-positive cells (infected with rcVSV-CoV2-S) in the different serum concentrations from 12 different individuals, with GFP-counts from the “no serum” control set to 100%. The researchers found variable responses against the different spike protein polymorphisms, and between individuals (Figure 3).
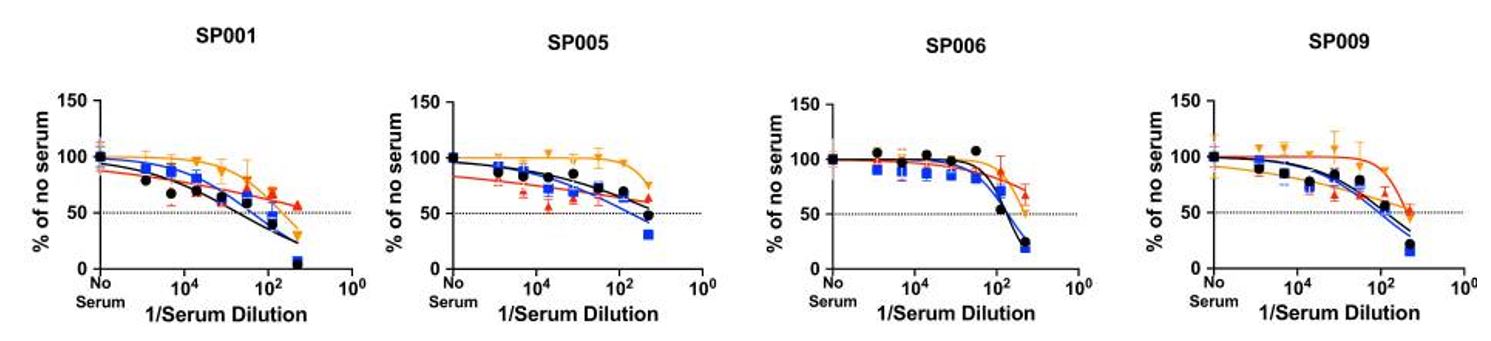
Figure 3: Representative data showing variable responses to SARS-CoV-2 variants. The microneutralization assay using rcVSV-CoV2-S was performed in 96-well microwell plates using virus stocks and serially diluted serum from vaccinated individuals. Imaging and quantification of GFP-positive, virus-infected cells were performed on Celigo. Figure and text adapted from Ikegame et al, 2021.
The speed of the Celigo Imaging Cytometer provides a significant advantage in virology workflows.
The Celigo’s integrated advanced analysis software and unique optics design enable the rapid readout of virology applications. For example, a neutralization assay (imaging and completed analysis) as described here by Ikegame et al, can be performed in ~2 minutes (Single channel Target 1 at 2um/pix) for a full 96 well plate. When combined with an automated plate stacker this complete workflow solution provides a throughput of at least 20 plates per hour.
Most imaging applications are performed in under 10 minutes, with the imaging and analysis (with complete data output) done in a few clicks, contrary to manual methods in which imaging and analysis can take hours (Figure 4).
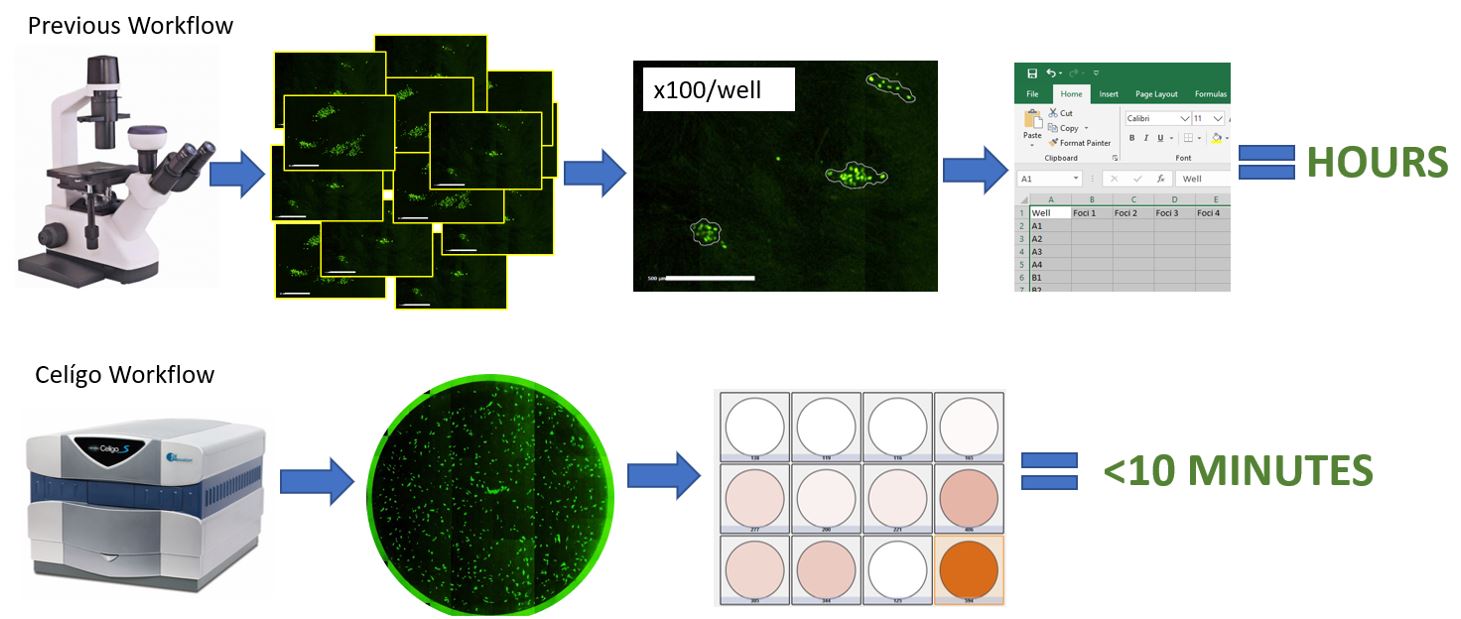
Figure 4: Celigo vs. Manual Imaging and Analysis. Celigo offers significant time savings.
In addition to the neutralization assays performed by Ikegame et al, the Celigo’s unique imaging technology has been routinely utilized for other common virology assays, including foci and plaque counting, antiviral screening, quantification of antiviral resistance to drugs, and cytopathic effect (CPE) quantification.
If you’d like to learn more about how Celigo can benefit your virology research, contact us for more information or request a live or virtual demonstration today!
References:
- Luan, Junwen, et al. “SARS-CoV-2 spike protein favors ACE2 from Bovidae and Cricetidae.” Journal of medical virology vol. 92,9 (2020): 1649-1656. doi:10.1002/jmv.25817
- Winger, Anna, and Thomas Caspari. “The Spike of Concern-The Novel Variants of SARS-CoV-2.” Viruses vol. 13,6 1002. 27 May. 2021, doi:10.3390/v13061002
- Logunov, Denis Y et al. “Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia.” Lancet (London, England) vol. 397,10275 (2021): 671-681. doi:10.1016/S0140-6736(21)00234-8
- Pearson, Morgan et al. “High-throughput viral microneutralization method for feline coronavirus using image cytometry.” Journal of virological methods vol. 286 (2020): 113979. doi:10.1016/j.jviromet.2020.113979


Leave A Comment